 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 103573 | ||||||||
| Common Name | MICPUN_103573 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Chlorophyta; prasinophytes; Mamiellophyceae; Mamiellales; Mamiellaceae; Micromonas
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 263aa MW: 28111 Da PI: 9.6675 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 29.5 | 1.6e-09 | 95 | 135 | 2 | 42 |
XXXCHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 2 kelkrerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeN 42
+e k+ rr+++NRe+ArrsRqRK+a++ eL ++L + +
103573 95 EEEKKMRRMISNRESARRSRQRKQARLSELSAATQNLWQDR 135
6889**************************98777776555 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.20.5.170 | 2.1E-7 | 90 | 163 | No hit | No description |
| SMART | SM00338 | 5.7E-7 | 94 | 158 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF00170 | 1.0E-6 | 95 | 131 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 9.381 | 96 | 159 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 1.58E-7 | 98 | 150 | No hit | No description |
| CDD | cd14702 | 2.01E-13 | 99 | 149 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 101 | 116 | IPR004827 | Basic-leucine zipper domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0046982 | Molecular Function | protein heterodimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 263 aa Download sequence Send to blast |
MVAERRATRA SAGCGFPDAN PVVKLESSSP KGKPPPTRAK TQAKPAAPPK LPAAKANVVG 60 PNASPPQFPC FAKEEADDLA LLETVPEEEG EEEDEEEKKM RRMISNRESA RRSRQRKQAR 120 LSELSAATQN LWQDRCQALE NVRMMTQLLV KARDENSRLE QELAVLGRHA LSMTDEGAVL 180 LRDLDQHAKI ASARQEASRE LRRYAKKLSR SPSPTIAASI MTAGGSDATA MTTAPVIVAA 240 PTAVAASGPV RVNLGAVSCP GA* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 110 | 116 | RRSRQRK |
| 2 | 110 | 117 | RRSRQRKQ |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00267 | DAP | Transfer from AT2G18160 | Download |
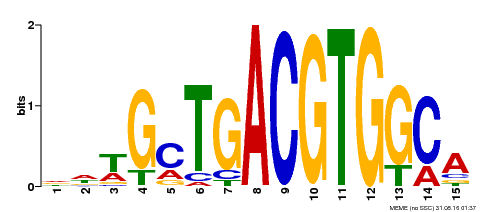
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | CP001577 | 0.0 | CP001577.1 Micromonas sp. RCC299 chromosome 12, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002509201.1 | 0.0 | predicted protein | ||||
| TrEMBL | C1FIP6 | 0.0 | C1FIP6_MICCC; Uncharacterized protein | ||||
| STRING | XP_002509201.1 | 0.0 | (Micromonas sp. RCC299) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Chlorophytae | OGCP15251 | 2 | 2 |
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | 103573 |
| Entrez Gene | 8247838 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



