 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 192902 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Chlorophyta; prasinophytes; Mamiellophyceae; Mamiellales; Mamiellaceae; Micromonas; Micromonas pusilla
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 655aa MW: 67391.4 Da PI: 10.3674 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 54.4 | 2.8e-17 | 25 | 71 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g WT++Ed++l +av+ + g++Wk+Ia+++ Rt+ qc +rwqk+l
192902 25 KGGWTPDEDDILRRAVAHYKGKNWKKIAEYFE-ERTDVQCLHRWQKVL 71
688*****************************.************986 PP
| |||||||
| 2 | Myb_DNA-binding | 62.6 | 8e-20 | 77 | 123 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT+eEd++++++vk lG ++W++Ia+ ++ gR +kqc++rw+++l
192902 77 KGPWTKEEDDKIIELVKALGAKQWSKIAQQLP-GRIGKQCRERWYNHL 123
79******************************.*************97 PP
| |||||||
| 3 | Myb_DNA-binding | 55.1 | 1.8e-17 | 129 | 173 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
r W+ eEd++l+ a++++G++ W+ Ia+t+ gRt++ +k++w++
192902 129 REEWSREEDQKLIVAHAEYGNR-WAEIAKTFV-GRTDNAIKNHWNST 173
679*******************.*********.***********986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 18.161 | 20 | 71 | IPR017930 | Myb domain |
| SMART | SM00717 | 2.8E-15 | 24 | 73 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.4E-15 | 25 | 71 | IPR001005 | SANT/Myb domain |
| SuperFamily | SSF46689 | 1.79E-15 | 26 | 81 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 4.9E-23 | 27 | 82 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 9.54E-14 | 28 | 71 | No hit | No description |
| PROSITE profile | PS51294 | 31.913 | 72 | 127 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 3.59E-30 | 74 | 170 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.2E-18 | 76 | 125 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.1E-18 | 77 | 123 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.46E-16 | 79 | 123 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 6.2E-27 | 83 | 126 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 2.5E-23 | 127 | 178 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 2.2E-16 | 128 | 176 | IPR001005 | SANT/Myb domain |
| PROSITE profile | PS51294 | 23.166 | 128 | 178 | IPR017930 | Myb domain |
| Pfam | PF00249 | 9.8E-15 | 129 | 173 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.00E-12 | 131 | 174 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 655 aa Download sequence Send to blast |
MLADKVTSLP PVVGRTGGPT RRSAKGGWTP DEDDILRRAV AHYKGKNWKK IAEYFEERTD 60 VQCLHRWQKV LNPELVKGPW TKEEDDKIIE LVKALGAKQW SKIAQQLPGR IGKQCRERWY 120 NHLNPEIKRE EWSREEDQKL IVAHAEYGNR WAEIAKTFVG RTDNAIKNHW NSTLKRKVDE 180 AVARGLPPLA AADAAADAEG KKAKNADKNG AKKQKAIAGA KASGAKASDV GGAAKTSKGK 240 KRKASDELAF GGPGANKRAN DPDIAHAAAQ MMGQYNLHAN GQGGNHAAAA AAAAAAAAAV 300 FDATLHNMLM SPTNGNSNGA AGAGAAPSPF GTGTTAAVQQ LFAASPSLYN NANSNGAPSP 360 DGKSGRPSPF PMTELFESMH DGSPTGKSNG AASPLGALST MMATGASPLL GNFAGGSPMN 420 LYNALLSSPG GSILRKNTVM RSGATPASNG ANANSMAPPR TQGGGEKTAG GNPEDASPSI 480 SGFGWFDTPG RGGAAAAKAG RTPAAKEPTP GRTTRASAAT NAAAAGPPRM ANAELAGLAS 540 PPHVKGGGAG IPGRLSSIFR RAPSGNDLKT ANAMKGGKEP AASDDVLEVM RQLEESNAGL 600 YMQAEAVMAR EAAEEEKRGG RATRAHGGVG GEDKGSPGAG GGNKFSPSQY LTAM* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1h88_C | 1e-67 | 24 | 178 | 5 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| 1h89_C | 1e-67 | 24 | 178 | 5 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00482 | DAP | Transfer from AT5G02320 | Download |
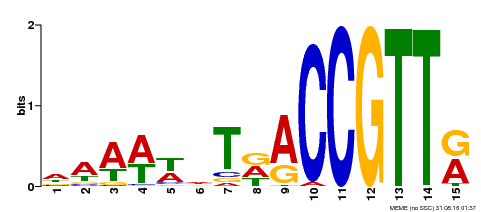
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_003057209.1 | 0.0 | predicted protein | ||||
| TrEMBL | C1MP22 | 0.0 | C1MP22_MICPC; Predicted protein | ||||
| STRING | XP_003057209.1 | 0.0 | (Micromonas pusilla) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Chlorophytae | OGCP15 | 16 | 114 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G02320.2 | 1e-84 | myb domain protein 3r-5 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | 192902 |



