 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 25644 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Chlorophyta; prasinophytes; Mamiellophyceae; Mamiellales; Bathycoccaceae; Ostreococcus
|
||||||||
| Family | GATA | ||||||||
| Protein Properties | Length: 386aa MW: 43066.9 Da PI: 6.9143 | ||||||||
| Description | GATA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | GATA | 33.1 | 7.8e-11 | 185 | 217 | 4 | 36 |
GATA 4 CgttkTplWRrgpdgnktLCnaCGlyyrkkglk 36
C +kTpl+Rrgpdg tLCnaCGl + ++ ++
25644 185 CDCSKTPLMRRGPDGIGTLCNACGLWWSRHQTM 217
67889************************9886 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00401 | 1.2E-6 | 174 | 229 | IPR000679 | Zinc finger, GATA-type |
| SuperFamily | SSF57716 | 6.18E-9 | 178 | 221 | No hit | No description |
| Gene3D | G3DSA:3.30.50.10 | 2.3E-12 | 178 | 219 | IPR013088 | Zinc finger, NHR/GATA-type |
| CDD | cd00202 | 2.88E-11 | 179 | 218 | No hit | No description |
| PROSITE pattern | PS00344 | 0 | 180 | 207 | IPR000679 | Zinc finger, GATA-type |
| Pfam | PF00320 | 5.5E-8 | 180 | 216 | IPR000679 | Zinc finger, GATA-type |
| PROSITE profile | PS50114 | 10.449 | 185 | 236 | IPR000679 | Zinc finger, GATA-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 386 aa Download sequence Send to blast |
MRAEPLDATA SNASASSPRE TRKRAREAIF ARFDALATAR GAKLAKRRGK RNCVELELEA 60 LGGVSDETLA KFATYGDKVQ KMMQKATPRG ATPGLAPIRY AARGTLFNEP YRVSVDEQGV 120 HRVVWGKEGE SRRVVRSKDG ARTPQEAVEC VHVEILRRMN ENIASVSDES HDGTFEARIC 180 RNCLCDCSKT PLMRRGPDGI GTLCNACGLW WSRHQTMREY PSVVPEETPH KAIFIRNPVK 240 SRRALKTLDV FGYYSSSVQA TLAKACAAVL QEERRSLRLP RVKSKPDVRY DFKRAIHDVF 300 DRCDCVFADS PSHSDDLGWE HYPPSITDFA AEQLEELEGV KLEGVKLEGV KLEGVELEGV 360 ELEELELEEL EAFAFGFEPP RLDPV* |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00369 | DAP | Transfer from AT3G21175 | Download |
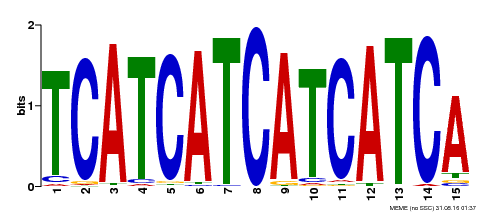
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | CP000596 | 0.0 | CP000596.1 Ostreococcus lucimarinus CCE9901 chromosome 16, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_001421825.1 | 0.0 | predicted protein | ||||
| TrEMBL | A4S8V3 | 0.0 | A4S8V3_OSTLU; Uncharacterized protein | ||||
| STRING | ABP00119 | 0.0 | (Ostreococcus 'lucimarinus') | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Chlorophytae | OGCP8134 | 5 | 5 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G21175.2 | 1e-14 | ZIM-like 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | 25644 |



