 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 28752.m000339 | ||||||||
| Common Name | LOC8264035, RCOM_0328910 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Euphorbiaceae; Acalyphoideae; Acalypheae; Ricinus
|
||||||||
| Family | AP2 | ||||||||
| Protein Properties | Length: 499aa MW: 55788.6 Da PI: 7.1013 | ||||||||
| Description | AP2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 53.6 | 5.6e-17 | 165 | 214 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkr.krfslgkfgtaeeAakaaiaarkkleg 55
s+y+GV++++++grW+++I+d + k+++lg f+ta Aa+a+++a+ k++g
28752.m000339 165 SQYRGVTFYRRTGRWESHIWD------CgKQVYLGGFDTAHAAARAYDRAAIKFRG 214
78*******************......55************************997 PP
| |||||||
| 2 | AP2 | 41.1 | 4.4e-13 | 257 | 307 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
s+y+GV+ +k grW+A+ + +k+++lg f+++ eAa+a+++a+ k +g
28752.m000339 257 SKYRGVTLHK-CGRWEARMGQF--L-GKKYIYLGLFDSEVEAARAYDKAAIKCNG 307
89********.7******5553..2.26**********99**********99776 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF00847 | 3.3E-9 | 165 | 214 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 1.24E-15 | 165 | 222 | IPR016177 | DNA-binding domain |
| CDD | cd00018 | 5.73E-11 | 166 | 221 | No hit | No description |
| PROSITE profile | PS51032 | 17.754 | 166 | 222 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 1.3E-31 | 166 | 228 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 8.7E-17 | 166 | 222 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 5.83E-26 | 257 | 317 | No hit | No description |
| Pfam | PF00847 | 1.3E-8 | 257 | 307 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 8.5E-17 | 257 | 316 | IPR016177 | DNA-binding domain |
| PROSITE profile | PS51032 | 15.527 | 258 | 315 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 2.5E-32 | 258 | 321 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 1.1E-15 | 258 | 315 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 5.5E-6 | 259 | 270 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 5.5E-6 | 297 | 317 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0055114 | Biological Process | oxidation-reduction process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0032440 | Molecular Function | 2-alkenal reductase [NAD(P)] activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 499 aa Download sequence Send to blast |
MLDLNLNVES SESTQNVDSV TLMEKYQYQY QYQYQHQQYN YTEGSGTTSN SSIANADASS 60 NDESCSTRAY TGGDTISTTT DSNNKNKIYT FNFDILKVGN EENENGAVIG TKELFPVENG 120 HGIDLSFERQ QQQQEIIGVG GEVRVMQQPL QQVKKSRRGP RSRSSQYRGV TFYRRTGRWE 180 SHIWDCGKQV YLGGFDTAHA AARAYDRAAI KFRGIDADIN FNLGDYDEDL KQMKNLTKEE 240 FVHILRRHST GFSRGSSKYR GVTLHKCGRW EARMGQFLGK KYIYLGLFDS EVEAARAYDK 300 AAIKCNGREA VTNFEPSTYE GEMISKASSE GSEHNLDLNL GISPSFGNCL KENEGHQQLH 360 SGPYDVYSKS SRENATSAMV GDPPFKGPLR TSDHPMFWNG IYPNFFPHEE RATDKRIELG 420 SSQGHPNWAW QMHGQVTATP MTMFSTAASS GFSFSATPPS AAIFPSKPLN PAAHNLFFSP 480 HATGGSQFYR PVKPPQAPP |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probably acts as a transcriptional activator. Binds to the GCC-box pathogenesis-related promoter element. May be involved in the regulation of gene expression by stress factors and by components of stress signal transduction pathways (By similarity). Regulates negatively the transition to flowering time and confers flowering time delay. {ECO:0000250, ECO:0000269|PubMed:14555699}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00615 | PBM | Transfer from AT2G28550 | Download |
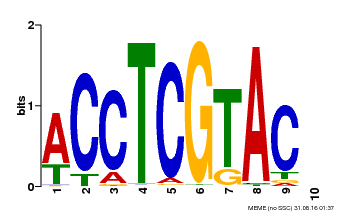
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 28752.m000339 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Repressed by miR172a-2/EAT. {ECO:0000269|PubMed:14555699}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002530247.1 | 0.0 | ethylene-responsive transcription factor RAP2-7 isoform X2 | ||||
| Swissprot | Q9SK03 | 1e-114 | RAP27_ARATH; Ethylene-responsive transcription factor RAP2-7 | ||||
| TrEMBL | B9SW78 | 0.0 | B9SW78_RICCO; Protein AINTEGUMENTA, putative | ||||
| STRING | XP_002530247.1 | 0.0 | (Ricinus communis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF2807 | 34 | 76 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G28550.3 | 1e-118 | related to AP2.7 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | 28752.m000339 |
| Entrez Gene | 8264035 |



