 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 28872.m000252 | ||||||||
| Common Name | RCOM_0351490 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Euphorbiaceae; Acalyphoideae; Acalypheae; Ricinus
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 389aa MW: 43627.9 Da PI: 5.2158 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 55.9 | 1.5e-17 | 34 | 80 | 1 | 48 |
NAM 1 lppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLp 48
l+pGfrFhPtdeelv++yLk+kv++k++++ ++i+evdiyk ePw+L
28872.m000252 34 LAPGFRFHPTDEELVSYYLKRKVSNKPVRF-NAISEVDIYKNEPWELA 80
579************************999.89*************96 PP
| |||||||
| 2 | NAM | 35.1 | 4e-11 | 81 | 108 | 101 | 128 |
NAM 101 glkktLvfykgrapkgektdWvmheyrl 128
+kktLvf++grap g++t+Wvmheyrl
28872.m000252 81 AMKKTLVFHSGRAPGGQRTNWVMHEYRL 108
69************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51005 | 17.058 | 1 | 132 | IPR003441 | NAC domain |
| SuperFamily | SSF101941 | 6.02E-33 | 30 | 133 | IPR003441 | NAC domain |
| Pfam | PF02365 | 3.2E-15 | 36 | 108 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009555 | Biological Process | pollen development | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0005730 | Cellular Component | nucleolus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 389 aa Download sequence Send to blast |
MGRETCTPPH APAPPSAAAV GAIQPAGAVK TTALAPGFRF HPTDEELVSY YLKRKVSNKP 60 VRFNAISEVD IYKNEPWELA AMKKTLVFHS GRAPGGQRTN WVMHEYRLVE EELDRLGALH 120 TDSYVLCRVF HKNNIGPPNG NRYAPFLEEE WDDGETALVP GEDAADEVVV GHDNTEMNRV 180 EQDNHSIRRD PLNINELPKD SQTVDEFRRH DLPVCKTERM DDCPPCVLNT EASFPLLQYK 240 RRKHNDEMVS NRSNASENST RTTQDPCSST TSTGATTTTT DTTMTTATAT TTAVSALLEF 300 SLMESIEPKE SPRVPTLKFD SFGLDSTVPP SCMKFINDLQ SEIHKISVER ETLKLEMMSA 360 QAMINILQSR IDFLNKENED LKRSIRCNQ |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional repressor that binds to the motif 5'-(C/T)A(C/A)G-3' in the promoter of target genes (PubMed:25578968). Binds also to the 5'-CTTGNNNNNCAAG-3' consensus sequence in chromatin (PubMed:26617990). Can bind to the mitochondrial dysfunction motif (MDM) present in the upstream regions of mitochondrial dysfunction stimulon (MDS) genes involved in mitochondrial retrograde regulation (MRR) (PubMed:24045019). Together with NAC050 and JMJ14, regulates gene expression and flowering time by associating with the histone demethylase JMJ14, probably by the promotion of RNA-mediated gene silencing (PubMed:25578968, PubMed:26617990). Regulates siRNA-dependent post-transcriptional gene silencing (PTGS) through SGS3 expression modulation (PubMed:28207953). Required during pollen development (PubMed:19237690). {ECO:0000269|PubMed:19237690, ECO:0000269|PubMed:24045019, ECO:0000269|PubMed:25578968, ECO:0000269|PubMed:26617990, ECO:0000269|PubMed:28207953}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00612 | ChIP-seq | Transfer from AT3G10490 | Download |
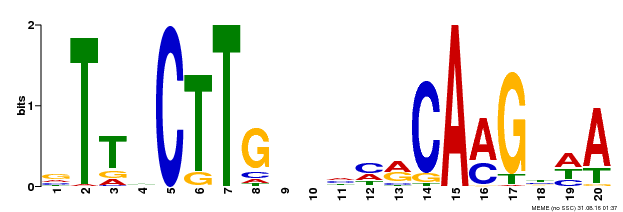
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 28872.m000252 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by type III effector proteins (TTEs) secreted by the pathogenic bacteria P.syringae pv. tomato DC3000 during basal defense. {ECO:0000269|PubMed:16553893}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015582996.1 | 0.0 | NAC domain containing protein 50 | ||||
| Swissprot | Q9SQY0 | 1e-111 | NAC52_ARATH; NAC domain containing protein 52 | ||||
| TrEMBL | B9T312 | 0.0 | B9T312_RICCO; Uncharacterized protein | ||||
| STRING | XP_002532631.1 | 0.0 | (Ricinus communis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF8189 | 32 | 41 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G10480.1 | 1e-104 | NAC domain containing protein 50 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | 28872.m000252 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



