 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 29596.m000720 | ||||||||
| Common Name | RCOM_0482900 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Euphorbiaceae; Acalyphoideae; Acalypheae; Ricinus
|
||||||||
| Family | GATA | ||||||||
| Protein Properties | Length: 324aa MW: 35525.2 Da PI: 7.919 | ||||||||
| Description | GATA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | GATA | 54 | 2.4e-17 | 210 | 246 | 1 | 35 |
GATA 1 CsnCgttk..TplWRrgpdgnktLCnaCGlyyrkkgl 35
C++Cgt++ Tp++Rrgp+g+++LCnaCGl+++ +g+
29596.m000720 210 CTHCGTSSksTPMMRRGPSGPRSLCNACGLFWANRGS 246
*********************************9986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00979 | 6.8E-11 | 74 | 109 | IPR010399 | Tify domain |
| PROSITE profile | PS51320 | 14.134 | 74 | 109 | IPR010399 | Tify domain |
| Pfam | PF06200 | 5.8E-12 | 78 | 108 | IPR010399 | Tify domain |
| Pfam | PF06203 | 1.4E-15 | 142 | 184 | IPR010402 | CCT domain |
| PROSITE profile | PS51017 | 12.739 | 142 | 184 | IPR010402 | CCT domain |
| SuperFamily | SSF57716 | 1.62E-10 | 203 | 246 | No hit | No description |
| SMART | SM00401 | 2.2E-14 | 204 | 258 | IPR000679 | Zinc finger, GATA-type |
| PROSITE profile | PS50114 | 9.249 | 204 | 245 | IPR000679 | Zinc finger, GATA-type |
| Gene3D | G3DSA:3.30.50.10 | 1.4E-14 | 208 | 245 | IPR013088 | Zinc finger, NHR/GATA-type |
| CDD | cd00202 | 6.98E-14 | 209 | 248 | No hit | No description |
| PROSITE pattern | PS00344 | 0 | 210 | 237 | IPR000679 | Zinc finger, GATA-type |
| Pfam | PF00320 | 4.8E-15 | 210 | 246 | IPR000679 | Zinc finger, GATA-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0007623 | Biological Process | circadian rhythm | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 324 aa Download sequence Send to blast |
MYSHSQSMNV HHQIHTPTGP TGDDEDGVAA DSIDHHHIRY EDGNATGVVL EDDVSHDSVY 60 VPTSAAGSEL AIQGNDVSSQ LTLTFRGQVY VFDAVTPDKV QAVLLLLGGC ELTSGPHGLE 120 VASQNQRSAV VDYPGRCTQP QRAASLNRFR QKRKERNFDK KVRYSVRQEV ALRMQRNKGQ 180 FTSSKKSDGT YGWGGGQDSG QDDSQQETSC THCGTSSKST PMMRRGPSGP RSLCNACGLF 240 WANRGSAMQR LNIFVGGSGV HFHALANALV WKNVHRQVME MNNVRGNFER SVQENPGSFS 300 ESNGAASNQR LKFQVVSQQQ PDQQ |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator that specifically binds 5'-GATA-3' or 5'-GAT-3' motifs within gene promoters. {ECO:0000250, ECO:0000269|PubMed:14966217}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00451 | DAP | Transfer from AT4G24470 | Download |
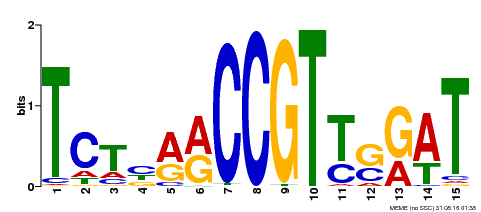
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 29596.m000720 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015579795.1 | 0.0 | GATA transcription factor 25 | ||||
| Swissprot | Q9LRH6 | 9e-82 | GAT25_ARATH; GATA transcription factor 25 | ||||
| TrEMBL | B9SN01 | 0.0 | B9SN01_RICCO; GATA transcription factor, putative | ||||
| STRING | XP_002527370.1 | 0.0 | (Ricinus communis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF2836 | 33 | 68 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G24470.2 | 6e-72 | GATA-type zinc finger protein with TIFY domain | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | 29596.m000720 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



