 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 29827.m002636 | ||||||||
| Common Name | RCOM_0867240 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Euphorbiaceae; Acalyphoideae; Acalypheae; Ricinus
|
||||||||
| Family | BES1 | ||||||||
| Protein Properties | Length: 668aa MW: 75470.6 Da PI: 6.0943 | ||||||||
| Description | BES1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF822 | 153.8 | 1.2e-47 | 70 | 211 | 2 | 143 |
DUF822 2 gsgrkptwkErEnnkrRERrRRaiaakiyaGLRaqGnyklpkraDnneVlkALcreAGwvvedDGttyrkgskpleeaeaagssasaspesslqss 97
g g+++++kE+E++k+RER+RRai+++++aGLR++Gn++lp+raD+n+Vl+AL+reAGw+ve DGttyr++ p + gs+ +s+es+++++
29827.m002636 70 GRGKREREKEKERTKLRERHRRAITSRMLAGLRQYGNFPLPARADMNDVLAALAREAGWTVESDGTTYRQSPAPS----QLGSFGVRSVESPVSTA 161
7899*****************************************************************888887....89999999999999988 PP
DUF822 98 lkssalaspvesysaspksssfpspssldsislasa....asllpvlsvl 143
+ al+ ++++++++ ++++sp slds+ +++ ++ p++sv+
29827.m002636 162 KAAAALECHNHHQQSVLRIDESLSPPSLDSVVMTEGdtrtDKFAPLTSVD 211
9999***************************9988765555555555554 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF05687 | 2.4E-48 | 70 | 213 | IPR008540 | BES1/BZR1 plant transcription factor, N-terminal |
| SuperFamily | SSF51445 | 4.45E-154 | 232 | 664 | IPR017853 | Glycoside hydrolase superfamily |
| Gene3D | G3DSA:3.20.20.80 | 5.4E-169 | 233 | 666 | IPR013781 | Glycoside hydrolase, catalytic domain |
| Pfam | PF01373 | 2.4E-79 | 258 | 626 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 6.5E-52 | 271 | 285 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 6.5E-52 | 292 | 310 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 6.5E-52 | 314 | 335 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 6.5E-52 | 407 | 429 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 6.5E-52 | 480 | 499 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 6.5E-52 | 514 | 530 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 6.5E-52 | 531 | 542 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 6.5E-52 | 549 | 572 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 6.5E-52 | 587 | 609 | IPR001554 | Glycoside hydrolase, family 14 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000272 | Biological Process | polysaccharide catabolic process | ||||
| GO:0048831 | Biological Process | regulation of shoot system development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0016161 | Molecular Function | beta-amylase activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 668 aa Download sequence Send to blast |
MTSNSNQDVL LDPQIDHYSQ TQSQPQPHPV QSNSHHIQPQ PRRPRGFAAT AAAAAAATDP 60 TNTTAVNVSG RGKREREKEK ERTKLRERHR RAITSRMLAG LRQYGNFPLP ARADMNDVLA 120 ALAREAGWTV ESDGTTYRQS PAPSQLGSFG VRSVESPVST AKAAAALECH NHHQQSVLRI 180 DESLSPPSLD SVVMTEGDTR TDKFAPLTSV DSLDADQLIQ DVRSGEHEGD FTSTSYVPVY 240 VMLATGFINN FCQLVDPQGV RQELSHIKSL DVDGVVVECW WGIVEAWGPQ KYVWSGYREL 300 FNIIREFKLK LQVVMAFYEY QGSDSEEVLI SLPQWVLEIG KENQDIFFTD REGRRNTECL 360 SWGIDKERVL KGRTGIEVYF DFMRSFRVEF DDLFAEGIIS AVEIGLGASG ELKYPCFPER 420 MGWRYPGIGE FQCYDKYLQQ NLRSAAQSRG HPFWARGPDN AGQYNSRPHE TGFFCERGDY 480 DSYFGRFFLH WYARTLIDHA DNVLSLASLT FEDTRIIVKI PAVYWWYKTS SHAAELTAGY 540 HNPTNQDGYS PVFEALKKHS VTVKFVCSGL QVSAHENDEV LADPEGLSWQ VLNSAWDRGL 600 TVAGVNVLSC YDREGCMRVV EMAKPRCNPD HRQFAFFVYQ QPSPLVPGTL CFTELDYFIK 660 CMHGKNKP |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wdq_A | 1e-121 | 235 | 626 | 10 | 401 | Beta-amylase |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00543 | DAP | Transfer from AT5G45300 | Download |
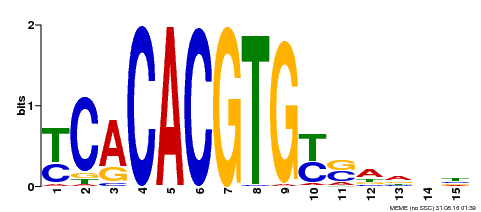
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 29827.m002636 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002519919.2 | 0.0 | beta-amylase 8 | ||||
| Swissprot | Q9FH80 | 0.0 | BAM8_ARATH; Beta-amylase 8 | ||||
| TrEMBL | B9S1Q4 | 0.0 | B9S1Q4_RICCO; Beta-amylase | ||||
| STRING | XP_002519919.1 | 0.0 | (Ricinus communis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF11902 | 30 | 32 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G45300.1 | 0.0 | beta-amylase 2 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | 29827.m002636 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



