 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 29889.m003258 | ||||||||
| Common Name | RCOM_0999660 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Euphorbiaceae; Acalyphoideae; Acalypheae; Ricinus
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 331aa MW: 36765.9 Da PI: 5.2477 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 48.8 | 1.7e-15 | 14 | 61 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+grWT+eEde+l +++ G g+W++ ++ g+ R++k+c++rw +yl
29889.m003258 14 KGRWTAEEDEILTKYILANGEGSWRSLPKNAGLLRCGKSCRLRWINYL 61
79******************************99************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 20.483 | 9 | 65 | IPR017930 | Myb domain |
| SMART | SM00717 | 5.6E-11 | 13 | 63 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 3.3E-13 | 14 | 61 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 4.0E-21 | 15 | 68 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 6.74E-20 | 15 | 88 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 1.18E-7 | 16 | 61 | No hit | No description |
| PROSITE profile | PS50090 | 4.031 | 62 | 121 | IPR017877 | Myb-like domain |
| SMART | SM00717 | 49 | 66 | 131 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.5E-7 | 69 | 124 | IPR009057 | Homeodomain-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 331 aa Download sequence Send to blast |
MGRAPCCEKV GLKKGRWTAE EDEILTKYIL ANGEGSWRSL PKNAGLLRCG KSCRLRWINY 60 LRADLKRGNI TKEEDETIVK LHTALGNRKI YSFSKNDSLS ITTINIADVP GACKSRSSNK 120 ITARSTIKRQ KQDRPKSGTL APDHEPAAKE KSITSSFVGQ VEGNQEMGYV QKGLKEHEPK 180 ENTRRESCMS SSGERFNATE VVCSRKEGES EVWGPYEWLD SEINRLKYAL EEGEVVGPSG 240 NVTNAIDDNT LQKDRESDVL GPSNPEGRGN EVMLGLDIVT SGDLERESSC SATAWSSNGE 300 SNGEWTQGIV DDDSRKELWD EGDKILSWLW G |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00668 | SELEX | Transfer from GRMZM2G016020 | Download |
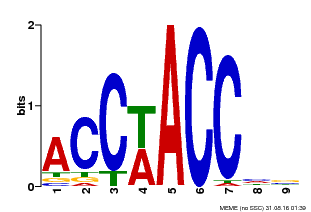
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 29889.m003258 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015574695.1 | 0.0 | transcription factor MYB111 | ||||
| TrEMBL | B9RZL8 | 0.0 | B9RZL8_RICCO; R2r3-myb transcription factor, putative | ||||
| STRING | XP_002519187.1 | 0.0 | (Ricinus communis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF31 | 34 | 817 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G49330.1 | 2e-49 | myb domain protein 111 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | 29889.m003258 |



