 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 29889.m003269 | ||||||||
| Common Name | RCOM_0999870 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Euphorbiaceae; Acalyphoideae; Acalypheae; Ricinus
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 918aa MW: 104145 Da PI: 7.4574 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 126.2 | 1.3e-39 | 31 | 136 | 3 | 118 |
CG-1 3 kekkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptfqrrc 98
+ ++rwl+++ei+aiL n++ ++++ ++ + p +k ++frkDG++wkkkkdgkt++E+he+LKvg+ e +++yYah+e+n+tf rrc
29889.m003269 31 EATSRWLRPNEIHAILCNYKYFTIHVKPVKLP----------RKAKNFRKDGHNWKKKKDGKTIKEAHEHLKVGNEERIHVYYAHGEDNSTFVRRC 116
4489*******************888777666..........67889************************************************* PP
CG-1 99 ywlLeeelekivlvhylevk 118
ywlL+++le+ivlvhy+e++
29889.m003269 117 YWLLDKTLEHIVLVHYRETQ 136
*****************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 62.658 | 25 | 141 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 5.2E-59 | 28 | 136 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 4.9E-34 | 31 | 134 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF01833 | 3.0E-4 | 369 | 455 | IPR002909 | IPT domain |
| SuperFamily | SSF81296 | 1.05E-12 | 370 | 456 | IPR014756 | Immunoglobulin E-set |
| CDD | cd00204 | 2.86E-15 | 553 | 663 | No hit | No description |
| Gene3D | G3DSA:1.25.40.20 | 5.9E-16 | 553 | 666 | IPR020683 | Ankyrin repeat-containing domain |
| Pfam | PF12796 | 4.0E-7 | 554 | 633 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 3.11E-17 | 556 | 668 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 15.592 | 571 | 636 | IPR020683 | Ankyrin repeat-containing domain |
| SMART | SM00248 | 1.7E-6 | 604 | 633 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 12.155 | 604 | 636 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF52540 | 3.19E-6 | 716 | 819 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| SMART | SM00015 | 34 | 749 | 771 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 7.712 | 769 | 796 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.011 | 771 | 789 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 190 | 772 | 790 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.0056 | 791 | 813 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 10.438 | 792 | 816 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.0053 | 794 | 813 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 31 | 871 | 893 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 7.895 | 873 | 901 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 918 aa Download sequence Send to blast |
MESSLPGRLV GSDIHGFHTL QDLDFGNIMA EATSRWLRPN EIHAILCNYK YFTIHVKPVK 60 LPRKAKNFRK DGHNWKKKKD GKTIKEAHEH LKVGNEERIH VYYAHGEDNS TFVRRCYWLL 120 DKTLEHIVLV HYRETQELQG SPVTPLNSNS SSVSDQSPRL LSEADSGTYV SDEKELQGDS 180 LTVINHELRL HEINTLEWDE LVTNDPNNSA TAKEGDGLSI ICYKIMGFAQ QNQIAVNGSM 240 NNGRYLSPYN LSAEISPLDN LTKPVVRSND SHFSIPDNEY IQSTGVQVNS NVQQKGSNFL 300 GTGDTLDMLV NDGLQSQDSF GRWIDYIIAD SPGSVDNAVL ESSFSSGLDS STSPAIDQLQ 360 SSVPEQIFVI TDISPAWAFS TETTKILVVG YFHEQYLQLA KSNMFCVCGD AYALVDIVQT 420 GVYRCLVSPH FPGIVNLFLS LDGHKPISQL INFEYRAPLH DPVVSSEDKT NWEEFKLQMR 480 LAHLLFSTSK SLGIQTSKVS SITLKEAKKF DHKTSNIHRS WAYLIKLIED NRLSFSQAKD 540 SLFELTLKSM LKEWLLERVV EGCKTTEYDA QGQGVIHLCS ILGYTWAVYL FSWSGLSLDF 600 RDKHGWTALH WAAYYGREKM VAVLLSAGAK PNLVTDPTKE NPDGCMAADL ASMKGYDGLA 660 AYLSEKALVA HFKDMSIAGN ASGTLQQTSA TDIVNSENLS EEELYLKDTL AAYRTAADAA 720 ARIQSAFREH SLKVRTTAVQ SANPEDEART IVAAMKIQHA YRNFETRKKM AAAVRIQYRF 780 RTWKMRKEFL NMRRQVIRIQ AAFRGYQVRR QYRKIIWSVG VLEKAILRWR LKRKGFRGLQ 840 IDPVEAVADL KQGSDTEEDF YKASRKQAEE RVERAVVRVQ AMFRSKKAQA EYRRMKLTHY 900 QVKLEYEELL DHDIDIDR |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator (PubMed:14581622). Binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Binds calmodulin in a calcium-dependent manner (By similarity). Involved in response to cold. Contributes together with CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:28351986). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:14581622, ECO:0000269|PubMed:28351986, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00435 | DAP | Transfer from AT4G16150 | Download |
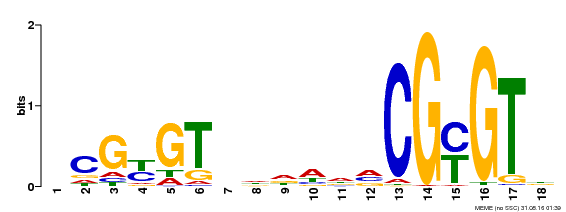
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 29889.m003269 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By heat shock, UVB, wounding, abscisic acid, H(2)O(2) and salicylic acid. {ECO:0000269|PubMed:12218065}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025013123.1 | 0.0 | LOW QUALITY PROTEIN: calmodulin-binding transcription activator 5 | ||||
| Swissprot | O23463 | 0.0 | CMTA5_ARATH; Calmodulin-binding transcription activator 5 | ||||
| TrEMBL | B9RZM9 | 0.0 | B9RZM9_RICCO; Calmodulin-binding transcription activator (Camta), plants, putative | ||||
| STRING | XP_002519198.1 | 0.0 | (Ricinus communis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF4288 | 34 | 59 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16150.1 | 0.0 | calmodulin binding;transcription regulators | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | 29889.m003269 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



