 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 29974.m000235 | ||||||||
| Common Name | LOC8271823, RCOM_1158820 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Euphorbiaceae; Acalyphoideae; Acalypheae; Ricinus
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 333aa MW: 37497 Da PI: 8.2876 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 177.5 | 3.7e-55 | 5 | 131 | 1 | 129 |
NAM 1 lppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLpkkvkaeekewyfFskrdkkyatgkrknratksgyWkatgkdkevlskk 96
lppGfrFhPtdeel+++yL++kv++ +++ ++i +vd++k+ePwdLp+k++ +ekewyfF rd+ky+tg r+nrat++gyWk+tgkdke+++
29974.m000235 5 LPPGFRFHPTDEELITCYLTRKVSDVRFTS-KAIVDVDLNKCEPWDLPAKASMGEKEWYFFNLRDRKYPTGLRTNRATEAGYWKTTGKDKEIFR-A 98
79*************************999.88***************999999****************************************.9 PP
NAM 97 gelvglkktLvfykgrapkgektdWvmheyrle 129
g lvg+kktLvfykgrap+gek++Wvmheyrle
29974.m000235 99 GILVGMKKTLVFYKGRAPRGEKSNWVMHEYRLE 131
99*****************************85 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 7.32E-63 | 3 | 151 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 59.333 | 5 | 151 | IPR003441 | NAC domain |
| Pfam | PF02365 | 4.8E-28 | 6 | 130 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 333 aa Download sequence Send to blast |
MEESLPPGFR FHPTDEELIT CYLTRKVSDV RFTSKAIVDV DLNKCEPWDL PAKASMGEKE 60 WYFFNLRDRK YPTGLRTNRA TEAGYWKTTG KDKEIFRAGI LVGMKKTLVF YKGRAPRGEK 120 SNWVMHEYRL ENKHLFKPTK EEWVVCRVFQ KSSSAAKKPQ QASSSQQSLG SPCDTNSVVN 180 EFGDIELPNL NSFVNSSSSG FNNMSAQSYN NENNVNMNMI NTNLNVNWAA TREAVVAAAT 240 LPSLSWPSSL LTSDLTMNSL ILKALQLRSY QQREAAAASN NDYSFLAQGI MPQFGSTDLS 300 SNFQASSSSS KGLESLPQQQ QQQEQPFNLD SIW |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ut4_A | 1e-57 | 4 | 157 | 16 | 171 | NO APICAL MERISTEM PROTEIN |
| 1ut4_B | 1e-57 | 4 | 157 | 16 | 171 | NO APICAL MERISTEM PROTEIN |
| 1ut7_A | 1e-57 | 4 | 157 | 16 | 171 | NO APICAL MERISTEM PROTEIN |
| 1ut7_B | 1e-57 | 4 | 157 | 16 | 171 | NO APICAL MERISTEM PROTEIN |
| 4dul_A | 1e-57 | 4 | 157 | 16 | 171 | NAC domain-containing protein 19 |
| 4dul_B | 1e-57 | 4 | 157 | 16 | 171 | NAC domain-containing protein 19 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Binds to the promoter regions of genes involved in chlorophyll catabolic processes, such as NYC1, SGR1, SGR2 and PAO. {ECO:0000269|PubMed:27021284}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00364 | DAP | Transfer from AT3G18400 | Download |
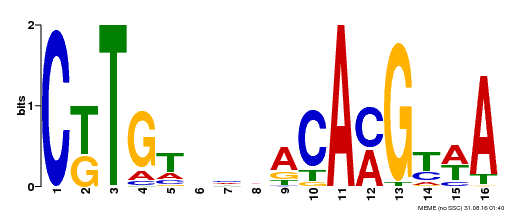
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 29974.m000235 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Repressed by the microRNA miR164. {ECO:0000269|PubMed:15294871, ECO:0000269|PubMed:17098808}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002532152.1 | 0.0 | NAC domain-containing protein 92 | ||||
| Swissprot | Q9FLJ2 | 4e-82 | NC100_ARATH; NAC domain-containing protein 100 | ||||
| TrEMBL | B9T1N3 | 0.0 | B9T1N3_RICCO; Transcription factor, putative | ||||
| STRING | XP_002532152.1 | 0.0 | (Ricinus communis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF4946 | 33 | 56 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G18400.1 | 1e-102 | NAC domain containing protein 58 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | 29974.m000235 |
| Entrez Gene | 8271823 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



