 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 30063.m001397 | ||||||||
| Common Name | RCOM_1296350 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Euphorbiaceae; Acalyphoideae; Acalypheae; Ricinus
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 1019aa MW: 114055 Da PI: 6.3905 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 71.6 | 1.1e-22 | 8 | 50 | 35 | 77 |
CG-1 35 ksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvgg 77
gsl+L++rk++ryfrkDG++w+kkkdgktvrE+hekLKv
30063.m001397 8 AGGSLFLFDRKVLRYFRKDGHNWRKKKDGKTVREAHEKLKVVC 50
579*************************************964 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 27.169 | 1 | 111 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 8.2E-7 | 2 | 75 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 3.4E-16 | 9 | 49 | IPR005559 | CG-1 DNA-binding domain |
| Gene3D | G3DSA:2.60.40.10 | 1.1E-6 | 414 | 519 | IPR013783 | Immunoglobulin-like fold |
| Pfam | PF01833 | 5.4E-6 | 433 | 511 | IPR002909 | IPT domain |
| SuperFamily | SSF81296 | 4.2E-17 | 433 | 518 | IPR014756 | Immunoglobulin E-set |
| Pfam | PF12796 | 2.6E-6 | 614 | 693 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 18.935 | 616 | 727 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 2.64E-17 | 617 | 727 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 1.0E-17 | 622 | 728 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 1.46E-12 | 622 | 725 | No hit | No description |
| SMART | SM00248 | 6.1E-4 | 666 | 695 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 11.087 | 666 | 698 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF52540 | 1.54E-7 | 841 | 892 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| SMART | SM00015 | 0.48 | 841 | 863 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 7.821 | 843 | 871 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.0015 | 844 | 862 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.023 | 864 | 886 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.395 | 865 | 889 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 4.5E-4 | 867 | 886 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0071275 | Biological Process | cellular response to aluminum ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1019 aa Download sequence Send to blast |
MALTHFLAGG SLFLFDRKVL RYFRKDGHNW RKKKDGKTVR EAHEKLKVVC LYNSEMQHSS 60 SNYRLEVLMY YTVIMPMVKR MRAFKGAVIG CLNSRRIAGL VVPDLTTSNS EIFLDILEGK 120 ERNGLQIVLL KFPLGNATES RLAYSWTNNS PSNSLIASSN KEPSGNTDST SPTSTLTSFC 180 EDADSADSQQ VNSGHHSYLE SPLMQSSPVI GKMNAGALSS HFLHPGSGPI SYVHGDRPGN 240 NGSSITEAQS LKSWEEVLEQ CTRGDKNAPS HLSTTSTQSD ATGIGRSMTL KLAYDLDTRL 300 LDQRTHNVDF PTTLEEFFSG PIHQNEELVH NNHHMLLTHA DQQLLMHTKS ENDMSVEENG 360 KYAFILKQPL LDGEEGLKKV DSFSRWVTRE LGEVDDLHMK SSSGIPWSTV ECGTVVDESS 420 LSPSLSQDQL FSIIDFSPKW GYADSKTEVH ISGTFLKSQH EVTKYNWSCM FGELEVPAEV 480 LADGILCCYA PPHSVASVPF YVTCSNRLAC SEVREFDYQS GSAEDVDVLD VYGGDAHDMY 540 LHLRLERLLS LRSSSPSCLF DGAREKHNLV EKLILLKEED EGCQVAETTS ERQLSQDEIR 600 NKFLQKGMQE KLYSWLLHTV AECGKGPSML DDDGQGMLHL AAALGYDWAI KPTMTAGVSI 660 NFRDVNGWTA LHWAAFYGRE QTVAALVSLG ADTRVLTDPS PEFPLGSTPA DLASGNGHKG 720 ISGFLAESSL TSYLHLLTLN DSVEGGAPEG SGMTAVQTIS ERMATPVKDG DVPNVLSLKD 780 SLTAIRNATQ AANRIYQVFR MQSFQRKQLT EYSDDEIGML DERALALIAA KTPKPLHSDG 840 VVNAAAIQIQ KKYRGWKKRK EFLIIRQRIV KIQAHIRGHQ VRKQYRTIIW SVGILEKVIL 900 RWRRKGSGLR GFRREALPIP KESNVQCENP KEDDYDFLKE GRKQNEVRQQ KALTRVKSMY 960 HCSEGQAQYR RLLNYFEKFR ETKENEMILS SPNEMAYGEN LIDIDSLLDD DTFMSIAFE |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Binds calmodulin in a calcium-dependent manner (By similarity). Involved in freezing tolerance in association with CAMTA1 and CAMTA3. Contributes together with CAMTA1 and CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:23581962). Involved together with CAMTA3 and CAMTA4 in the positive regulation of a general stress response (PubMed:25039701). Involved in tolerance to aluminum. Binds to the promoter of ALMT1 transporter and contributes to the positive regulation of aluminum-induced expression of ALMT1 (PubMed:25627216). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:23581962, ECO:0000269|PubMed:25039701, ECO:0000269|PubMed:25627216, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00043 | PBM | Transfer from AT5G64220 | Download |
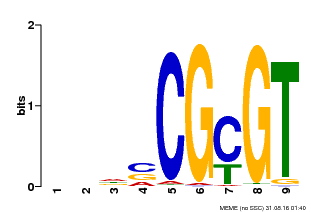
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 30063.m001397 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By salt, wounding, abscisic acid, H(2)O(2) and salicylic acid (PubMed:12218065). Induced by aluminum (PubMed:25627216). {ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:25627216}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AP007304 | 5e-32 | AP007304.2 Lotus japonicus genomic DNA, chromosome 4, clone: LjB10F10, BM1099, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015574793.1 | 0.0 | calmodulin-binding transcription activator 2 | ||||
| Swissprot | Q6NPP4 | 0.0 | CMTA2_ARATH; Calmodulin-binding transcription activator 2 | ||||
| TrEMBL | B9S036 | 0.0 | B9S036_RICCO; Calmodulin-binding transcription activator (Camta), plants, putative | ||||
| STRING | XP_002519355.1 | 0.0 | (Ricinus communis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF7406 | 26 | 42 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G64220.2 | 0.0 | Calmodulin-binding transcription activator protein with CG-1 and Ankyrin domains | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | 30063.m001397 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



