 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 30068.m002661 | ||||||||
| Common Name | LOC8278522, RCOM_1316780 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Euphorbiaceae; Acalyphoideae; Acalypheae; Ricinus
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 405aa MW: 43100.3 Da PI: 6.5258 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 70.3 | 2.9e-22 | 301 | 363 | 1 | 63 |
XXXXCHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 1 ekelkrerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelkkevaklksev 63
e+elkr+rrkq+NRe+ArrsR+RK+ae++eL++++++L++eN L++e++++k e+++l +e+
30068.m002661 301 ERELKRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENANLRSEVNRIKSEYEQLLAEN 363
89*********************************************************9987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF07777 | 2.3E-30 | 1 | 97 | IPR012900 | G-box binding protein, multifunctional mosaic region |
| Pfam | PF16596 | 4.8E-55 | 136 | 268 | No hit | No description |
| Gene3D | G3DSA:1.20.5.170 | 2.6E-17 | 294 | 359 | No hit | No description |
| SMART | SM00338 | 5.1E-21 | 301 | 365 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF00170 | 4.3E-20 | 301 | 363 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 13.334 | 303 | 366 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 1.06E-10 | 304 | 360 | No hit | No description |
| CDD | cd14702 | 2.15E-24 | 306 | 356 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 308 | 323 | IPR004827 | Basic-leucine zipper domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 405 aa Download sequence Send to blast |
MGSSDMDKTA KEKESKTQPP TTTQEQSSAT TTGTVNPDWT GFQAYSPIPP HGFVASSPQA 60 HPYMWGVQPI MPPYGTPPHP YVAMYPHSGI YAHPSIPPGS YPFSPFAMPS PNGIAEASGY 120 TPGNTEPDGK PSDVKEKLPI KRSKGSLGSL NMITGKNNEL GKTSGASANG AYSKSAESGS 180 EGTSEGSDAN SQNDSQMKSG GRQDSEDASQ NGGSAHGLQN GGQANTVMNQ TMSIVPISAT 240 GAPGALPGPA TNLNIGMDYW GATSSAIPAI RGKVPSTPVA GGVVTPGSRD AVQSQLWLQD 300 ERELKRQRRK QSNRESARRS RLRKQAECDE LAQRAEALKE ENANLRSEVN RIKSEYEQLL 360 AENASLKERL GEIPGNDDLR ASRNDQHLSN DTQKTEQTEI VQAGH |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 317 | 323 | RRSRLRK |
| 2 | 317 | 324 | RRSRLRKQ |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator that binds to the G-box motif (5'-CACGTG-3') and other cis-acting elements with 5'-ACGT-3' core, such as Hex, C-box and as-1 motifs. Possesses high binding affinity to G-box, much lower affinity to Hex and C-box, and little affinity to as-1 element (PubMed:18315949). G-box and G-box-like motifs are cis-acting elements defined in promoters of certain plant genes which are regulated by such diverse stimuli as light-induction or hormone control (Probable). Binds to the G-box motif 5'-CACGTG-3' of LHCB2.4 (At3g27690) promoter. May act as transcriptional repressor in light-regulated expression of LHCB2.4. Binds DNA as monomer. DNA-binding activity is redox-dependent (PubMed:22718771). {ECO:0000269|PubMed:18315949, ECO:0000269|PubMed:22718771, ECO:0000305|PubMed:18315949}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00291 | DAP | Transfer from AT2G35530 | Download |
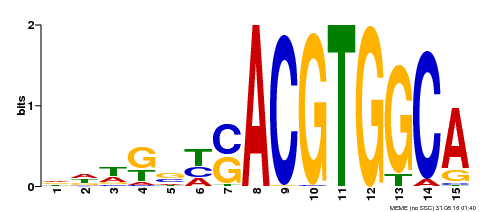
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 30068.m002661 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002518994.1 | 0.0 | bZIP transcription factor 16 | ||||
| Refseq | XP_025013063.1 | 0.0 | bZIP transcription factor 16 | ||||
| Refseq | XP_025013064.1 | 0.0 | bZIP transcription factor 16 | ||||
| Swissprot | Q501B2 | 1e-174 | BZP16_ARATH; bZIP transcription factor 16 | ||||
| TrEMBL | B9RZ25 | 0.0 | B9RZ25_RICCO; DNA-binding protein EMBP-1, putative | ||||
| STRING | XP_002518994.1 | 0.0 | (Ricinus communis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF4017 | 32 | 56 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G35530.1 | 1e-156 | basic region/leucine zipper transcription factor 16 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | 30068.m002661 |
| Entrez Gene | 8278522 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



