 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 30076.m004464 | ||||||||
| Common Name | LOC8270765, RCOM_1340450 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Euphorbiaceae; Acalyphoideae; Acalypheae; Ricinus
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 768aa MW: 83834.1 Da PI: 6.5479 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 50.5 | 4.7e-16 | 25 | 69 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
r rWT+eE+ ++++a k++G W +I +++g ++t+ q++s+ qk+
30076.m004464 25 RERWTEEEHNRFLEALKLYGRA-WQRIEEHIG-TKTAVQIRSHAQKF 69
78******************88.*********.************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 6.28E-17 | 19 | 75 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 21.091 | 20 | 74 | IPR017930 | Myb domain |
| TIGRFAMs | TIGR01557 | 8.4E-17 | 23 | 72 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 7.3E-13 | 24 | 72 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.6E-8 | 25 | 65 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 3.2E-13 | 25 | 68 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.49E-9 | 27 | 70 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009739 | Biological Process | response to gibberellin | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0010243 | Biological Process | response to organonitrogen compound | ||||
| GO:0042754 | Biological Process | negative regulation of circadian rhythm | ||||
| GO:0043496 | Biological Process | regulation of protein homodimerization activity | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0048574 | Biological Process | long-day photoperiodism, flowering | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 768 aa Download sequence Send to blast |
MDTYSSGGED LVIKTRKPYT ITKQRERWTE EEHNRFLEAL KLYGRAWQRI EEHIGTKTAV 60 QIRSHAQKFF SKLEKEAIAK GVPIGQALDI DIPPPRPKRK PSNPYPRKTG AGPTPSQVAA 120 KDGKLPSLVS FPRCTQVLDL EKEPLPERLN GHEKQTDATE NQGDNCSEVF TLLQEAHCSS 180 VSSANKNSVV TAEALKNSCS FREFVPSLKK VVNQDATNES YVTIELEGNQ KLDKPDAKQT 240 VQDNGSSKAS KSESCLFHEK FDQAKKSDEF NSALPTDEME TMQGYPRHVP VHVLEGSLGT 300 CMQTPTSDVS FQEPIFCPTG EVHGHPNLYS HPAASATTEH QNTAPRSSTH QSFPSFPPFN 360 STQHNQDDYR SFLHISSTFS SLIVSTLLQN PAAHSAASFA ATFWPYANLE SSADSPICPQ 420 GGFPSRQMNS APSMAAIAAA TVAAATAWWA AHGLLPLCAP LHAAFTCPPA SGTAVASTGA 480 GQVPAAKTER KLTVENPLLQ NQQFDVEHSK VLQAQNSASK SLEMSLSDSE ESGGPKKNTG 540 SKATDHEMAT PAPEVQDPSK AKARKPADRS SCGSNTSSSS EVETDALEKL EKGNEELKET 600 DTNPEPTESS CRRSRSNSSI SDSWKEVSEE GRLAFQALFS REVLPQSFSP PHVLKNEARQ 660 KDEIEEDKQN TVEKNENALL LSLNGNISGF CTSHQEAEKI EMPRCENNGE DGLLTFGLGH 720 GKLKARRTGF KPYKRCSVEA KENRMLTAGS QGEEKGPKRI RVEGKAST |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00103 | PBM | Transfer from AT2G46830 | Download |
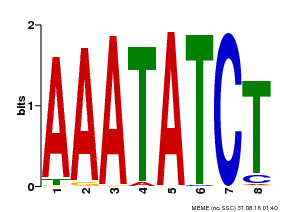
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 30076.m004464 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002515093.1 | 0.0 | protein LHY isoform X1 | ||||
| Refseq | XP_015572367.1 | 0.0 | protein LHY isoform X1 | ||||
| TrEMBL | B9RMV4 | 0.0 | B9RMV4_RICCO; Uncharacterized protein | ||||
| STRING | XP_002515093.1 | 0.0 | (Ricinus communis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF4860 | 25 | 37 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G46830.1 | 1e-81 | circadian clock associated 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | 30076.m004464 |
| Entrez Gene | 8270765 |



