 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 30190.m011120 | ||||||||
| Common Name | LOC8265404, RCOM_1683570 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Euphorbiaceae; Acalyphoideae; Acalypheae; Ricinus
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 230aa MW: 25995 Da PI: 6.1715 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 51.8 | 2e-16 | 57 | 107 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
+ ++GVr ++++ +Wv+e+r+p +k+ r++lg++ t+e+Aa+a++ a+++l+g
30190.m011120 57 PVFRGVR-NRNNDKWVCELREP---NKKSRIWLGTYPTPEMAARAHDVAALALRG 107
579***9.888**********8...347*************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF00847 | 1.6E-11 | 57 | 107 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 6.2E-27 | 58 | 121 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 3.86E-20 | 58 | 117 | IPR016177 | DNA-binding domain |
| PROSITE profile | PS51032 | 20.916 | 58 | 115 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 9.8E-30 | 58 | 117 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 2.4E-6 | 59 | 70 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 3.00E-27 | 59 | 117 | No hit | No description |
| PRINTS | PR00367 | 2.4E-6 | 81 | 97 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010371 | Biological Process | regulation of gibberellin biosynthetic process | ||||
| GO:0016049 | Biological Process | cell growth | ||||
| GO:0048510 | Biological Process | regulation of timing of transition from vegetative to reproductive phase | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 230 aa Download sequence Send to blast |
MQTQFSDPYP FDKHESFSPS DSTTSRRAVH SDEEVLLATS YPKKRAGRRV FKETRHPVFR 60 GVRNRNNDKW VCELREPNKK SRIWLGTYPT PEMAARAHDV AALALRGKSA CLNFADSAWR 120 LPVPVSTDPN DIRRAAHEAA ELFRPQEFGG QVTRKFGSVV EQDAGEASSS DSKQEVSYID 180 EEDFLDMPGL LARMAERPLL SPPHYARNDG MELGDHLEAD IDVSLWSYSV |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
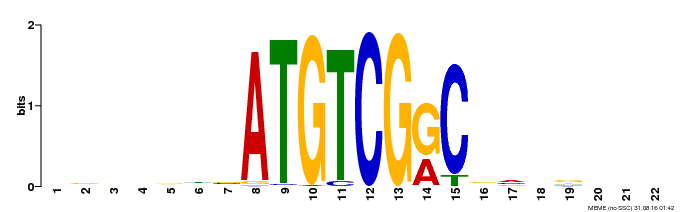
| UniProt | Transcriptional activator that binds specifically to the DNA sequence 5'-[AG]CCGAC-3'. Binding to the C-repeat/DRE element mediates cold or dehydration-inducible transcription. CBF/DREB1 factors play a key role in freezing tolerance and cold acclimation. {ECO:0000269|PubMed:11798174}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00002 | PBM | Transfer from 475045 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 30190.m011120 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002509703.1 | 1e-172 | dehydration-responsive element-binding protein 1E | ||||
| Swissprot | Q9SGJ6 | 4e-67 | DRE1E_ARATH; Dehydration-responsive element-binding protein 1E | ||||
| TrEMBL | B9RC19 | 1e-170 | B9RC19_RICCO; Dehydration-responsive element-binding protein 1F, putative | ||||
| STRING | XP_002509703.1 | 1e-171 | (Ricinus communis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF243 | 33 | 224 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G63030.2 | 5e-59 | ERF family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | 30190.m011120 |
| Entrez Gene | 8265404 |



