 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 312456 | ||||||||
| Common Name | ARALYDRAFT_312456 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Arabidopsis
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 495aa MW: 54006 Da PI: 9.0239 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 14.4 | 0.00011 | 66 | 86 | 3 | 23 |
ETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 3 CpdCgksFsrksnLkrHirtH 23
C+ C+k F r nL+ H r H
312456 66 CEVCNKGFQREQNLQLHRRGH 86
*******************88 PP
| |||||||
| 2 | zf-C2H2 | 12.8 | 0.00036 | 140 | 162 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirtH 23
+kC +C+k++ +s+ k H +t+
312456 140 WKCDKCSKRYAVQSDWKAHSKTC 162
58*****************9998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00355 | 0.025 | 64 | 86 | IPR015880 | Zinc finger, C2H2-like |
| SuperFamily | SSF57667 | 2.52E-7 | 64 | 86 | No hit | No description |
| PROSITE profile | PS50157 | 10.97 | 64 | 86 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 2.0E-5 | 64 | 86 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE pattern | PS00028 | 0 | 66 | 86 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 140 | 105 | 135 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 3.5E-5 | 128 | 161 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 2.52E-7 | 135 | 160 | No hit | No description |
| SMART | SM00355 | 140 | 140 | 160 | IPR015880 | Zinc finger, C2H2-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003676 | Molecular Function | nucleic acid binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 495 aa Download sequence Send to blast |
MSSSYNTIAS SSAQTFLLSG AASGARPNNF NREEAARTMI QQPNSSVTPP PKKRRNQPGN 60 PSKFLCEVCN KGFQREQNLQ LHRRGHNLPW KLKQKSNKEV RRKVYLCPEA SCVHHDPARA 120 LGDLTGIKKH YYRKHGEKKW KCDKCSKRYA VQSDWKAHSK TCGTKEYRCD CGTIFSSERE 180 KDSEGERKIK DAKFGHIGWF HCLINEYCGQ RDIVGVVDEH FTFLGRDSYI THRAFCDALI 240 QESARNPTVS FTAMAPAAGG GTRNGFYGGA SAALSHNHFG NNSNSGFTPL AAGYNLNRSS 300 SDKFEDFVPQ STNPNPGPTN FLMQCSPNQG LLAQNNQSLM NQHGLISLGD NTNHNLFNIG 360 YFQDTKNSDQ IGVPSLFTNG ADNNDPSAFL RGLTSSSSPT VVVNDFGDSD NGNLQGLMNS 420 LAATTDQQGR PTSLLDLHFG NNLSMGGADR LTLDFLGVNG GIVSTVNGRG GRSGGPPLDA 480 EMKFSHPTHP FGKA* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5b3h_C | 3e-24 | 136 | 250 | 3 | 69 | Zinc finger protein JACKDAW |
| 5b3h_F | 3e-24 | 136 | 250 | 3 | 69 | Zinc finger protein JACKDAW |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. {ECO:0000305}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00142 | DAP | Transfer from AT1G14580 | Download |
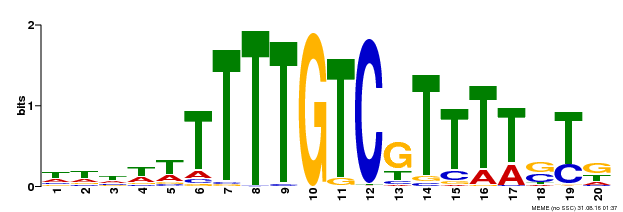
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 312456 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY091041 | 0.0 | AY091041.1 Arabidopsis thaliana putative zinc finger protein (At1g14580) mRNA, complete cds. | |||
| GenBank | AY133766 | 0.0 | AY133766.1 Arabidopsis thaliana clone U18944 putative zinc finger protein (At1g14580) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020869197.1 | 0.0 | protein indeterminate-domain 6, chloroplastic isoform X1 | ||||
| Refseq | XP_020869198.1 | 0.0 | protein indeterminate-domain 6, chloroplastic isoform X2 | ||||
| Refseq | XP_020869199.1 | 0.0 | protein indeterminate-domain 6, chloroplastic isoform X2 | ||||
| Swissprot | Q8RWX7 | 0.0 | IDD6_ARATH; Protein indeterminate-domain 6, chloroplastic | ||||
| TrEMBL | D7KBM3 | 0.0 | D7KBM3_ARALL; T5E21.8 | ||||
| STRING | fgenesh1_pm.C_scaffold_1001236 | 0.0 | (Arabidopsis lyrata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM3678 | 17 | 59 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G14580.2 | 0.0 | C2H2-like zinc finger protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | 312456 |
| Entrez Gene | 9326127 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



