 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 324546 | ||||||||
| Common Name | ARALYDRAFT_324546 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Arabidopsis
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 812aa MW: 88695.4 Da PI: 6.7182 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 62.2 | 7.7e-20 | 114 | 169 | 1 | 56 |
TT--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 1 rrkRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
+++ +++t++q++ Le++F+++ +p++++r +L+++l+L+ rqVk+WFqNrR+++k
324546 114 KKRYHRHTPKQIQDLESVFKECAHPDEKQRLDLSRRLNLDPRQVKFWFQNRRTQMK 169
688999***********************************************999 PP
| |||||||
| 2 | START | 181 | 6.8e-57 | 320 | 539 | 2 | 206 |
HHHHHHHHHHHHHHHC-TT-EEEE....EXCCTTEEEEEEESSS......SCEEEEEEEECCSCHHHHHHHHHCCCGGCT-TT-S....EEEEEEEECTT... CS
START 2 laeeaaqelvkkalaeepgWvkss....esengdevlqkfeeskv.....dsgealrasgvvdmvlallveellddkeqWdetla....kaetlevissg... 88
la++ ++elvk+a+ ++p+Wv+ss e++n++e+ ++f++ + + +ea+++ g v+ ++ lve+l+d+ +W e+++ + +t+e issg
324546 320 LALASMDELVKMAQTRDPLWVRSSdtgfEMLNQEEYDTSFTRCVGpkpdgYVSEASKEAGTVIINSLALVETLMDSE-RWAEMFPsmisRTSTTEIISSGmgg 421
68899***********************9999999999997755499999***************************.*******9999************** PP
...EEEEEEEEXXTTXX-SSX.EEEEEEEEEEE.TTS-EEEEEEEEE-TTS--.-TTSEE-EESSEEEEEEEECTCEEEEEEEE-EE--SSXXHHHHHHHHHH CS
START 89 ...galqlmvaelqalsplvp.RdfvfvRyirqlgagdwvivdvSvdseqkppesssvvRaellpSgiliepksnghskvtwvehvdlkgrlphwllrslvks 187
gal+lm aelq+lsplvp R + f+R+++q+ +g+w++vdvS+ds ++ + sss+ R lpSg+l+++++ng skvtw+eh++++++ +h l+r+l++
324546 422 srnGALHLMHAELQLLSPLVPvRQVSFLRFCKQHAEGVWAVVDVSIDSIREGS-SSSCRR---LPSGCLVQDMANGCSKVTWIEHTEYDENRIHRLYRPLLSC 520
****************************************************9.777766...**************************************** PP
HHHHHHHHHHHHTXXXXXX CS
START 188 glaegaktwvatlqrqcek 206
gla+ga +w+a+lqrqce+
324546 521 GLAFGAHRWMAALQRQCEC 539
*****************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 2.2E-20 | 88 | 172 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 2.92E-19 | 102 | 171 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.086 | 111 | 171 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 1.9E-18 | 112 | 175 | IPR001356 | Homeobox domain |
| Pfam | PF00046 | 1.8E-17 | 114 | 169 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 1.80E-17 | 114 | 171 | No hit | No description |
| PROSITE pattern | PS00027 | 0 | 146 | 169 | IPR017970 | Homeobox, conserved site |
| PROSITE profile | PS50848 | 39.403 | 310 | 542 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 2.75E-30 | 313 | 539 | No hit | No description |
| CDD | cd08875 | 2.45E-108 | 314 | 538 | No hit | No description |
| SMART | SM00234 | 9.8E-45 | 319 | 539 | IPR002913 | START domain |
| Pfam | PF01852 | 4.3E-49 | 320 | 539 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 2.47E-17 | 568 | 803 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0048497 | Biological Process | maintenance of floral organ identity | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 812 aa Download sequence Send to blast |
MNFNGFLDDG SPGASKLLSD IPYNNNHFSF SAVDTTMLGT TAIAPPHSRP FSSSGLSLGL 60 QTNGEMSRNG EIFESNVTRK SSRGEDVESR SESDNAEAVS GDDLDTSDRP LKKKKRYHRH 120 TPKQIQDLES VFKECAHPDE KQRLDLSRRL NLDPRQVKFW FQNRRTQMKT QIERHENALL 180 RQENDKLRAE NMSVREAMRN PMCGNCGGPA VIGEISMEEQ HLRIENSRLK DELDRVCALT 240 GKFLGRSNGS HHIPDSALVL GVGVGCNVGG GFTLSSPVLP QASPRFEISN ATGSGLVATV 300 NRQQPVSVSD FDQRSRYLDL ALASMDELVK MAQTRDPLWV RSSDTGFEML NQEEYDTSFT 360 RCVGPKPDGY VSEASKEAGT VIINSLALVE TLMDSERWAE MFPSMISRTS TTEIISSGMG 420 GSRNGALHLM HAELQLLSPL VPVRQVSFLR FCKQHAEGVW AVVDVSIDSI REGSSSSCRR 480 LPSGCLVQDM ANGCSKVTWI EHTEYDENRI HRLYRPLLSC GLAFGAHRWM AALQRQCECL 540 TILMSSTVSP SPNPTPINCN GRKSMLKLAK RMTDNFCGGV CASSLQKWSK LNVGNVDEDV 600 RIMTRKSVNN PGEPPGIILN AATSVWMPIS PRRLFDFLGN ERLRSEWDIL SNGGPMKEMA 660 HIAKGHDHSN SVSLLRASAI NANQSSMLIL QETSIDAAGA LVVYAPVDIP AMQAVMNGGD 720 SAYVALLPSG FAILPNAQAG TQRCAAEERN ANGNGNGGCM EEGGSLLTVA FQILVNSLPT 780 AKLTVESVET VNNLISCTVQ KIKAALHCDS T* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00418 | DAP | Transfer from AT3G61150 | Download |
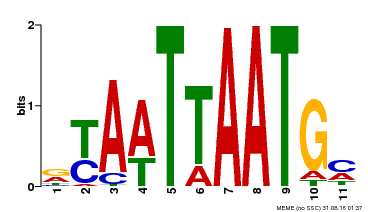
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 324546 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY050866 | 0.0 | AY050866.1 Arabidopsis thaliana putative homeobox protein (At3g61150) mRNA, complete cds. | |||
| GenBank | AY096757 | 0.0 | AY096757.1 Arabidopsis thaliana putative homeobox protein (At3g61150) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002876596.1 | 0.0 | homeobox-leucine zipper protein HDG1 isoform X1 | ||||
| Swissprot | Q9M2E8 | 0.0 | HDG1_ARATH; Homeobox-leucine zipper protein HDG1 | ||||
| TrEMBL | D7LS63 | 0.0 | D7LS63_ARALL; Uncharacterized protein | ||||
| STRING | fgenesh1_pm.C_scaffold_5002090 | 0.0 | (Arabidopsis lyrata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM1128 | 27 | 105 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G61150.1 | 0.0 | homeodomain GLABROUS 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | 324546 |
| Entrez Gene | 9312664 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



