 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 324904 | ||||||||
| Common Name | ARALYDRAFT_324904, MYB3R-5 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Arabidopsis
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 542aa MW: 60494.7 Da PI: 7.4307 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 55.3 | 1.6e-17 | 75 | 121 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g WT+eEde l +av ++ g++Wk+Ia+ ++ Rt+ qc +rwqk+l
324904 75 KGGWTPEEDETLRRAVDKYKGKRWKKIAEFFP-ERTEVQCLHRWQKVL 121
688*****************************.************986 PP
| |||||||
| 2 | Myb_DNA-binding | 61.9 | 1.3e-19 | 127 | 173 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT+eEd+++v++vk++G+ W+ Ia+ ++ gR +kqc++rw+++l
324904 127 KGPWTQEEDDKIVELVKKYGPAKWSVIAKSLP-GRIGKQCRERWHNHL 173
79******************************.*************97 PP
| |||||||
| 3 | Myb_DNA-binding | 46.6 | 8.1e-15 | 179 | 222 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqk 46
+++WT eE+ l + ++++G++ W+ Ia+ ++ gRt++ +k++w++
324904 179 KDAWTIEEESALMNSHRMYGNK-WAEIAKVLP-GRTDNAIKNHWNS 222
679*******************.*********.***********96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 18.518 | 70 | 121 | IPR017930 | Myb domain |
| SMART | SM00717 | 9.8E-16 | 74 | 123 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 5.4E-16 | 75 | 121 | IPR001005 | SANT/Myb domain |
| SuperFamily | SSF46689 | 7.18E-16 | 76 | 131 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 1.8E-23 | 77 | 129 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 6.80E-14 | 78 | 121 | No hit | No description |
| PROSITE profile | PS51294 | 31.882 | 122 | 177 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 7.34E-32 | 124 | 220 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 3.9E-18 | 126 | 175 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 2.3E-18 | 127 | 173 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.86E-15 | 129 | 173 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 7.7E-27 | 130 | 176 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 4.3E-22 | 177 | 228 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 21.069 | 178 | 228 | IPR017930 | Myb domain |
| SMART | SM00717 | 1.7E-13 | 178 | 226 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 2.8E-13 | 179 | 222 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 3.67E-10 | 181 | 224 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 542 aa Download sequence Send to blast |
MSSSSNPAAC SPEKEERSEL KIEIQCMENK QPLAASCSSA SEGSSCFFLK SPEIATPATV 60 SSTPRRTSGP MRRAKGGWTP EEDETLRRAV DKYKGKRWKK IAEFFPERTE VQCLHRWQKV 120 LNPELVKGPW TQEEDDKIVE LVKKYGPAKW SVIAKSLPGR IGKQCRERWH NHLNPGIRKD 180 AWTIEEESAL MNSHRMYGNK WAEIAKVLPG RTDNAIKNHW NSSLKKKLEF YLATGNLPPP 240 ATKFSVLNDI GDGDRDSKQS SATKPFKDSD SVTQTSSGNT DSNEVGRDHF DSSSALLEEV 300 AASRRIGVNE YACSPVEYKP QLPNLEPISE EVRINSKAYG ERSIQRKEEN GFGTPKHGSL 360 YYKSPLDYYF PSEADLQHMY GYECGCSPGA ATPVSLMTPP CNKDSGLAAT RSPESFLREA 420 ARTFPNTPSI FRKRRKVVLA AKADDDVVNV GVKEVDQKED SKDSSDISPA GRESLLLETS 480 DDCRDDDKLE PNGNAFNVSP PYRLRAKRTA VIKSRQLEFT FAKEKHETET SELASEEDKP 540 V* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1h88_C | 6e-69 | 74 | 228 | 5 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| 1h89_C | 6e-69 | 74 | 228 | 5 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that binds 5'-AACGG-3' motifs in gene promoters (By similarity). Transcription repressor that regulates organ growth. Binds to the promoters of G2/M-specific genes and to E2F target genes to prevent their expression in post-mitotic cells and to restrict the time window of their expression in proliferating cells (PubMed:26069325). {ECO:0000250|UniProtKB:Q94FL9, ECO:0000269|PubMed:26069325}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00482 | DAP | Transfer from AT5G02320 | Download |
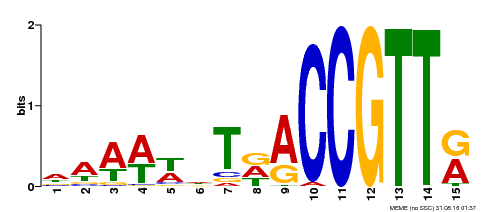
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 324904 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Slightly induced by ethylene and salicylic acid (SA). {ECO:0000269|PubMed:16463103}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY519649 | 0.0 | AY519649.1 Arabidopsis thaliana MYB transcription factor (At5g02320) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002873047.1 | 0.0 | transcription factor MYB3R-5 | ||||
| Swissprot | Q6R032 | 0.0 | MB3R5_ARATH; Transcription factor MYB3R-5 | ||||
| TrEMBL | D7M7S2 | 0.0 | D7M7S2_ARALL; Myb domain protein 3R-5 | ||||
| STRING | fgenesh1_pm.C_scaffold_6000124 | 0.0 | (Arabidopsis lyrata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM1667 | 28 | 87 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G02320.2 | 0.0 | myb domain protein 3r-5 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | 324904 |
| Entrez Gene | 9307039 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



