 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 341194 | ||||||||
| Common Name | ARALYDRAFT_341194 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Arabidopsis
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 286aa MW: 32496.2 Da PI: 6.1782 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 45.9 | 1.3e-14 | 94 | 138 | 3 | 47 |
SS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
+W++ E++l++d+ +++G+g+Wk+Iar+ k+R++ q+ s+ qky
341194 94 AWSPNEHKLFLDGLNKYGKGDWKSIARECVKTRSPMQVASHAQKY 138
6*******************************************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00717 | 1.3 | 3 | 54 | IPR001005 | SANT/Myb domain |
| SuperFamily | SSF46689 | 1.94E-6 | 6 | 60 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 14.215 | 87 | 143 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 2.62E-16 | 88 | 144 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 7.2E-10 | 89 | 146 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 5.3E-15 | 90 | 142 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 1.9E-8 | 91 | 141 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 2.2E-11 | 94 | 138 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 2.67E-10 | 94 | 139 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 286 aa Download sequence Send to blast |
MDEIPLWTRV DDKRFESALV QFPEGSPYFL ENIAQFLQKP LKDVKYYYQA LVDDVALIES 60 GNFALPNYRD DDYVSLKEAT KSKNQGTGKK KGIAWSPNEH KLFLDGLNKY GKGDWKSIAR 120 ECVKTRSPMQ VASHAQKYFL RKNKKGKRMS IHDMPLGDAD NVTVPVSHLN STGQQPQFGD 180 QILPDHYYHC SQDNVTNIPG SNLVFMGQQP HFGGQIPPNQ SHPYSRDNVT VPESNLKPMG 240 EQPHFGDHIS PDQYDRDFLD NFGFFYDDGE DDGSLASFEK LYYKA* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that coordinates abscisic acid (ABA) biosynthesis and signaling-related genes via binding to the specific promoter motif 5'-(A/T)AACCAT-3'. Represses ABA-mediated salt (e.g. NaCl and KCl) stress tolerance. Regulates leaf shape and promotes vegetative growth. {ECO:0000269|PubMed:26243618}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00344 | DAP | Transfer from AT3G10580 | Download |
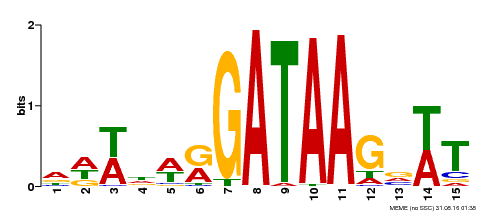
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 341194 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by salicylic acid (SA) and gibberellic acid (GA) (PubMed:16463103). Triggered by dehydration and salt stress (PubMed:26243618). {ECO:0000269|PubMed:16463103, ECO:0000269|PubMed:26243618}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AC011560 | 1e-157 | AC011560.6 Arabidopsis thaliana chromosome 3 BAC F13M14 genomic sequence, complete sequence. | |||
| GenBank | ATAC013428 | 1e-157 | AC013428.6 Arabidopsis thaliana chromosome III BAC F18K10 genomic sequence, complete sequence. | |||
| GenBank | CP002686 | 1e-157 | CP002686.1 Arabidopsis thaliana chromosome 3, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002884799.1 | 0.0 | transcription factor DIVARICATA | ||||
| Swissprot | Q9FNN6 | 8e-34 | SRM1_ARATH; Transcription factor SRM1 | ||||
| TrEMBL | D7L911 | 0.0 | D7L911_ARALL; Uncharacterized protein | ||||
| STRING | fgenesh1_pg.C_scaffold_3000957 | 0.0 | (Arabidopsis lyrata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM14731 | 7 | 19 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G10580.1 | 1e-141 | MYB_related family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | 341194 |
| Entrez Gene | 9318753 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



