 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 34891 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Chlorophyta; prasinophytes; Mamiellophyceae; Mamiellales; Bathycoccaceae; Ostreococcus
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 180aa MW: 20770.5 Da PI: 10.4966 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 57.5 | 3e-18 | 19 | 66 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g WT+eEdell av+ +Gg++Wk+Ia ++ Rt+ qc +rwqk+l
34891 19 KGGWTPEEDELLRGAVAVYGGRNWKKIAVYFSDSRTDVQCLHRWQKVL 66
688******************************************986 PP
| |||||||
| 2 | Myb_DNA-binding | 65.6 | 9.4e-21 | 72 | 118 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT+eEd +++++v++lG ++W++Ia +++ gR +kqc++rw+++l
34891 72 KGPWTAEEDARIIELVTELGAKRWSKIAGELP-GRIGKQCRERWYNHL 118
79******************************.*************97 PP
| |||||||
| 3 | Myb_DNA-binding | 53.9 | 4e-17 | 124 | 168 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
r W+++Ed +l+ a++q+G++ W+ Ia+ + gRt++ +k++w++
34891 124 REEWSADEDRQLIIAHAQYGNR-WAEIAKSFK-GRTDNAIKNHWNST 168
789*******************.********9.***********986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 17.34 | 14 | 66 | IPR017930 | Myb domain |
| SMART | SM00717 | 1.7E-14 | 18 | 68 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.2E-15 | 19 | 66 | IPR001005 | SANT/Myb domain |
| SuperFamily | SSF46689 | 9.31E-17 | 20 | 76 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 1.6E-22 | 21 | 78 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 9.65E-11 | 22 | 66 | No hit | No description |
| PROSITE profile | PS51294 | 31.45 | 67 | 122 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.13E-30 | 69 | 165 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 3.4E-17 | 71 | 120 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.3E-18 | 72 | 118 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 4.71E-14 | 74 | 118 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 2.7E-27 | 79 | 125 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 23.221 | 123 | 173 | IPR017930 | Myb domain |
| SMART | SM00717 | 4.0E-16 | 123 | 171 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 4.0E-15 | 124 | 168 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 4.53E-11 | 126 | 169 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 4.7E-23 | 126 | 173 | IPR009057 | Homeodomain-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0032875 | Biological Process | regulation of DNA endoreduplication | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003713 | Molecular Function | transcription coactivator activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 180 aa Download sequence Send to blast |
RRARVASRKI GGPTRRSAKG GWTPEEDELL RGAVAVYGGR NWKKIAVYFS DSRTDVQCLH 60 RWQKVLNPEL VKGPWTAEED ARIIELVTEL GAKRWSKIAG ELPGRIGKQC RERWYNHLDP 120 EIKREEWSAD EDRQLIIAHA QYGNRWAEIA KSFKGRTDNA IKNHWNSTLK RKVDQALNQG |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1h88_C | 1e-67 | 18 | 173 | 5 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| 1h89_C | 1e-67 | 18 | 173 | 5 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor involved in abiotic stress responses (PubMed:17293435, PubMed:19279197). May play a regulatory role in tolerance to salt, cold, and drought stresses (PubMed:17293435). Transcriptional activator that binds specifically to a mitosis-specific activator cis-element 5'-(T/C)C(T/C)AACGG(T/C)(T/C)A-3', found in promoters of cyclin genes such as CYCB1-1 and KNOLLE (AC Q84R43). Positively regulates a subset of G2/M phase-specific genes, including CYCB1-1, CYCB2-1, CYCB2-2, and CDC20.1 in response to cold treatment (PubMed:19279197). {ECO:0000269|PubMed:17293435, ECO:0000269|PubMed:19279197}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00505 | DAP | Transfer from AT5G11510 | Download |
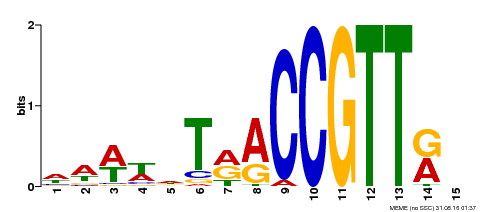
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by cold, drought and salt stresses. {ECO:0000269|PubMed:17293435}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | CP000589 | 0.0 | CP000589.1 Ostreococcus lucimarinus CCE9901 chromosome 9, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_001419697.1 | 1e-130 | predicted protein, partial | ||||
| Swissprot | Q0JHU7 | 2e-76 | MB3R2_ORYSJ; Transcription factor MYB3R-2 | ||||
| TrEMBL | A4S269 | 1e-128 | A4S269_OSTLU; Uncharacterized protein (Fragment) | ||||
| STRING | ABO97990 | 1e-129 | (Ostreococcus 'lucimarinus') | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Chlorophytae | OGCP15 | 16 | 114 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G02320.2 | 2e-78 | myb domain protein 3r-5 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | 34891 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



