 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 39026 | ||||||||
| Common Name | MICPUCDRAFT_39026 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Chlorophyta; prasinophytes; Mamiellophyceae; Mamiellales; Mamiellaceae; Micromonas; Micromonas pusilla
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 369aa MW: 38181.3 Da PI: 9.8164 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 51.9 | 1.7e-16 | 14 | 59 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
+++WT+eEde+l + ++ +G+g+W+++a+ + gR+ k+c++rw +
39026 14 KNSWTPEEDERLRERIAAHGPGNWSAVAAFLE-GRSSKSCRLRWCNQ 59
679*****************************.***********985 PP
| |||||||
| 2 | Myb_DNA-binding | 56.3 | 7.6e-18 | 66 | 110 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
rg++T+eEd+l++ a++++G++ W+ I+r ++ gRt++q+k+r+++
39026 66 RGPFTAEEDKLILAAHAMHGNK-WAIISRSIP-GRTDNQVKNRYNST 110
89********************.*********.***********986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 18.511 | 9 | 60 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.2E-30 | 12 | 107 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 3.3E-16 | 13 | 62 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 2.15E-14 | 16 | 58 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 9.1E-25 | 16 | 67 | IPR009057 | Homeodomain-like |
| Pfam | PF13921 | 4.6E-16 | 17 | 76 | No hit | No description |
| PROSITE profile | PS51294 | 26.057 | 61 | 115 | IPR017930 | Myb domain |
| SMART | SM00717 | 5.2E-17 | 65 | 113 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 2.00E-12 | 68 | 111 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 6.8E-24 | 68 | 113 | IPR009057 | Homeodomain-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0048527 | Biological Process | lateral root development | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 369 aa Download sequence Send to blast |
MTNVDEDGDD KMKKNSWTPE EDERLRERIA AHGPGNWSAV AAFLEGRSSK SCRLRWCNQL 60 NPDVKRGPFT AEEDKLILAA HAMHGNKWAI ISRSIPGRTD NQVKNRYNST LRRVSTTARR 120 ERDAVAKAPA KRKATNTEAA ASPGEAASPS AKRSKKPPPE RAPPTPTTVS PKRARAVSPS 180 SGSEVDETVT GLERLVQASF AVEREEGEER DSGFGIRDSG SDAATASAAV AAFDANANAT 240 VHGGFNIADQ ARIMMANIQM TAQLAQLAAM SRGAAPALSA PAGEGPGFGA TAAAATAAAA 300 PAFFGGGGGA AAAAASPPAS QNASTHGENH FLSHAHAQMM AATFAAAAAA DGGRRVLYTG 360 SHTTTSAW* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1h8a_C | 2e-32 | 12 | 113 | 25 | 126 | MYB TRANSFORMING PROTEIN |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00400 | DAP | Transfer from AT3G50060 | Download |
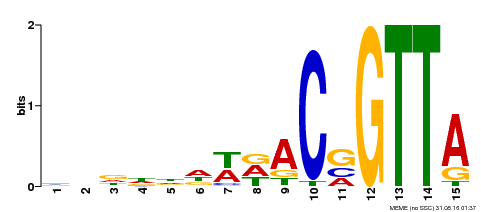
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_003057507.1 | 0.0 | predicted protein | ||||
| TrEMBL | C1MN26 | 0.0 | C1MN26_MICPC; Predicted protein | ||||
| STRING | XP_003057507.1 | 0.0 | (Micromonas pusilla) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Chlorophytae | OGCP15 | 16 | 114 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G67300.1 | 8e-38 | myb domain protein r1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | 39026 |
| Entrez Gene | 9682469 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



