 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 423935 | ||||||||
| Common Name | SELMODRAFT_423935 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Lycopodiidae; Selaginellales; Selaginellaceae; Selaginella
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 341aa MW: 38533 Da PI: 7.4429 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 104.4 | 6.7e-33 | 77 | 130 | 2 | 55 |
G2-like 2 prlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
prlrWtp+LHe+Fv+av++LGG+++AtPk +l+lm+vkgLt++hvkSHLQ+YR+
423935 77 PRLRWTPDLHECFVRAVDRLGGQDRATPKLVLQLMGVKGLTIAHVKSHLQMYRS 130
8****************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 12.316 | 73 | 133 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.5E-32 | 73 | 132 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 6.63E-16 | 75 | 133 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.3E-23 | 77 | 131 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 1.2E-9 | 78 | 129 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 341 aa Download sequence Send to blast |
MQLEVEMDQG ASSPPLHNSR KRLSSVEEDD LDAPGSSDSN NTVSGDGSSD GATVLGGPFG 60 SSSARSSVRQ YIRSKMPRLR WTPDLHECFV RAVDRLGGQD RATPKLVLQL MGVKGLTIAH 120 VKSHLQMYRS MKNDENGPVV MEERKGEQAQ AAVASDASLL HHPWTPQLQQ IRGEKKCRDN 180 PNAQQQYYAH NYFHRPVLQP LDCHARTEDA WSQIASPNQS QEWLSRLSIA ATKCYSERFN 240 DLQIGWRKNH HTLFQQTGGD DNTLRSRSLE REESRIHSSI LLSQNRINSC PALIHFDEIL 300 EGSRSAMKHH ENFNFHPQKL CYITDLCTKT TLEPRELTFY * |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4r_A | 4e-19 | 78 | 132 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 4e-19 | 78 | 132 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 4e-19 | 78 | 132 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 4e-19 | 78 | 132 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00297 | DAP | Transfer from AT2G38300 | Download |
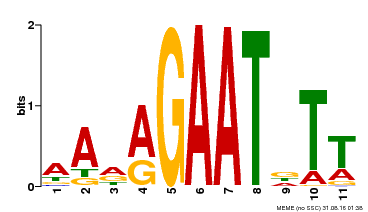
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024545718.1 | 0.0 | protein PHOSPHATE STARVATION RESPONSE 3 | ||||
| TrEMBL | D8SNA0 | 0.0 | D8SNA0_SELML; Uncharacterized protein | ||||
| STRING | EFJ14034 | 0.0 | (Selaginella moellendorffii) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Representative plant | OGRP285 | 15 | 123 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G38300.1 | 2e-33 | G2-like family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | 423935 |
| Entrez Gene | 9645119 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



