 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 462846124 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Chloridoideae; Eragrostideae; Eragrostidinae; Eragrostis
|
||||||||
| Family | Dof | ||||||||
| Protein Properties | Length: 311aa MW: 32490.1 Da PI: 9.9957 | ||||||||
| Description | Dof family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-Dof | 125.8 | 1.3e-39 | 13 | 73 | 2 | 62 |
zf-Dof 2 kekalkcprCdstntkfCyynnyslsqPryfCkaCrryWtkGGalrnvPvGggrrknkkss 62
+e a+kcprCdstntkfCyynnyslsqPr+fCk+CrryWt+GG+lrnvPvGgg+r+nk+ss
462846124 13 PEPAIKCPRCDSTNTKFCYYNNYSLSQPRHFCKTCRRYWTQGGSLRNVPVGGGCRRNKRSS 73
67899*****************************************************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| ProDom | PD007478 | 3.0E-33 | 1 | 71 | IPR003851 | Zinc finger, Dof-type |
| Pfam | PF02701 | 2.4E-33 | 16 | 71 | IPR003851 | Zinc finger, Dof-type |
| PROSITE profile | PS50884 | 29.599 | 17 | 71 | IPR003851 | Zinc finger, Dof-type |
| PROSITE pattern | PS01361 | 0 | 19 | 55 | IPR003851 | Zinc finger, Dof-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009640 | Biological Process | photomorphogenesis | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 311 aa Download sequence Send to blast |
MAERARLMRM PQPEPAIKCP RCDSTNTKFC YYNNYSLSQP RHFCKTCRRY WTQGGSLRNV 60 PVGGGCRRNK RSSKSSSSSS GASSSKPSSS SATRQLPGST TAGGGGAIAG GGIISPAGLG 120 SLSQQHHQYL PFLGSMHHQP NLGLAFPTGL PALGMQHVDA ADQFPVASGG ASLDQWRVHQ 180 QQPAQQQFPF LAAGGMLDLH QPRQQQQMYQ QLGLEAAANR GSSGAAAFTL GQASGATARQ 240 DGSMKLDDSK GQEMSLQRQY MAALRQGEGI WGGNAGGSGG ADGGGNGGVS WPMNIPGFRS 300 SSTGDDGTGL L |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00407 | DAP | Transfer from AT3G55370 | Download |
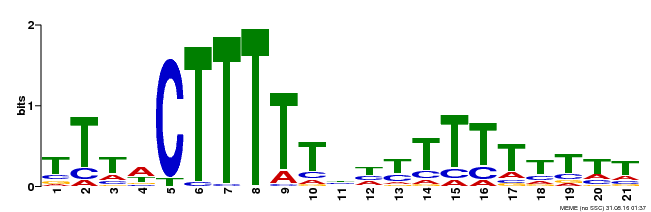
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 462846124 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | JN870914 | 2e-99 | JN870914.1 Sorghum bicolor dof protein (dof3) gene, partial cds. | |||
| GenBank | KC582400 | 2e-99 | KC582400.1 Sorghum bicolor dof3 protein (dof3) gene, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025791450.1 | 1e-105 | dof zinc finger protein DOF5.1-like isoform X1 | ||||
| TrEMBL | A0A3L6TN10 | 1e-104 | A0A3L6TN10_PANMI; Dof zinc finger protein DOF3.6 isoform X2 | ||||
| STRING | Pavir.J14669.1.p | 1e-104 | (Panicum virgatum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP12301 | 24 | 32 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G55370.1 | 1e-39 | OBF-binding protein 3 | ||||



