 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 462846634 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Chloridoideae; Eragrostideae; Eragrostidinae; Eragrostis
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 605aa MW: 69329.9 Da PI: 8.2594 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 13.7 | 0.00019 | 70 | 92 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirtH 23
+kC Cg +F + +Lk+H+++H
462846634 70 HKCLECGACFQKPAHLKQHMQSH 92
79*******************99 PP
| |||||||
| 2 | zf-C2H2 | 22.5 | 2.9e-07 | 151 | 175 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirtH 23
+ Cp dC+ s++rk++L+rH+ tH
462846634 151 FNCPleDCPFSYKRKDHLNRHMLTH 175
78**********************9 PP
| |||||||
| 3 | zf-C2H2 | 15.4 | 5.4e-05 | 179 | 204 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt.H 23
++C C+++Fs k n++rHi+ H
462846634 179 FSCAvdGCDRRFSIKANMQRHIKEiH 204
789999*****************988 PP
| |||||||
| 4 | zf-C2H2 | 17.1 | 1.5e-05 | 216 | 239 | 1 | 23 |
EEET.TTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCp.dCgksFsrksnLkrHirtH 23
++C+ C+ksF+ s Lk+H +H
462846634 216 FVCKeGCNKSFKYLSRLKKHEESH 239
89******************9988 PP
| |||||||
| 5 | zf-C2H2 | 15.6 | 4.5e-05 | 306 | 330 | 2 | 23 |
EET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 2 kCp..dCgksFsrksnLkrHirt.H 23
kC+ C +Fs+ksnL++H++ H
462846634 306 KCTfeGCEHTFSNKSNLTKHMKAcH 330
799999****************955 PP
| |||||||
| 6 | zf-C2H2 | 15.6 | 4.5e-05 | 486 | 510 | 2 | 23 |
EET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 2 kCp..dCgksFsrksnLkrHirt.H 23
kC+ C +Fs+ksnL++H++ H
462846634 486 KCTfeGCEHTFSNKSNLTKHMKAcH 510
799999****************955 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00355 | 23 | 24 | 47 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 11.593 | 70 | 97 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 1.8E-5 | 70 | 95 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.035 | 70 | 92 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 72 | 92 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 6.3E-7 | 149 | 177 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 3.1E-4 | 151 | 175 | IPR015880 | Zinc finger, C2H2-like |
| Pfam | PF00096 | 2.7E-5 | 151 | 175 | IPR007087 | Zinc finger, C2H2 |
| PROSITE profile | PS50157 | 10.097 | 151 | 180 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 153 | 175 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 1.03E-8 | 161 | 210 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 6.3E-8 | 179 | 206 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 11.468 | 179 | 209 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.0027 | 179 | 204 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 181 | 204 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 4.3E-5 | 212 | 239 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 10.575 | 216 | 239 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.13 | 216 | 239 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 218 | 239 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 8.2 | 247 | 272 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 249 | 272 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 15 | 275 | 296 | IPR015880 | Zinc finger, C2H2-like |
| SuperFamily | SSF57667 | 2.95E-8 | 290 | 347 | No hit | No description |
| PROSITE profile | PS50157 | 11.884 | 305 | 335 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.012 | 305 | 330 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 1.6E-6 | 306 | 330 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE pattern | PS00028 | 0 | 307 | 330 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 7.6E-5 | 331 | 358 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 5.9 | 336 | 362 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 338 | 362 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 8.2 | 427 | 452 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 429 | 452 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 15 | 455 | 476 | IPR015880 | Zinc finger, C2H2-like |
| SuperFamily | SSF57667 | 2.44E-8 | 470 | 527 | No hit | No description |
| PROSITE profile | PS50157 | 11.884 | 485 | 515 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.012 | 485 | 510 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 1.6E-6 | 486 | 510 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE pattern | PS00028 | 0 | 487 | 510 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 7.6E-5 | 511 | 538 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 5.9 | 516 | 542 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 518 | 542 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005730 | Cellular Component | nucleolus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008097 | Molecular Function | 5S rRNA binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| GO:0080084 | Molecular Function | 5S rDNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 605 aa Download sequence Send to blast |
MCSGDEIDGG GRVGQATCKD IRRYKCEFCA IVRSKKCLIQ AHMVENHKDE LDQSEIYNSN 60 GEKIIYEVEH KCLECGACFQ KPAHLKQHMQ SHSQEVGMTS CEASTRLPRS RQPIELPYGC 120 FEKMHEDPIS KCHLRKLGPF FISSQLAMRL FNCPLEDCPF SYKRKDHLNR HMLTHQGKFS 180 CAVDGCDRRF SIKANMQRHI KEIHEDENTA KSDQQFVCKE GCNKSFKYLS RLKKHEESHV 240 KLNYVEVVCC EPGCMKMFTN VECLRAHNQS CHQYIQCEIC GEKHLKKNIK RHLQAHEEVP 300 SSERMKCTFE GCEHTFSNKS NLTKHMKACH DNVKPFSCRF AGCDKAFTYK HVRDNHEQSS 360 AHGDFEEIDA QLQSRPRGGR KRKSLTVETL TRKRVTIPGQ ASATDDGVEY LRWLLSGGDV 420 KLNYVEVVCC EPGCMKMFTN VECLRAHNQS CHQYIQCEIC GEKHLKKNIK RHLQAHEEVP 480 SSERMKCTFE GCEHTFSNKS NLTKHMKACH DNVKPFSCRF AGCDKAFTYK HVRDNHEQSS 540 AHGDFEEIDA QLQSRPRGGR KRKALTVETL TRKRVTIPGE ASATDDGVGY LRWLLSGGDG 600 SSQNL |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1tf6_A | 2e-12 | 151 | 332 | 43 | 190 | PROTEIN (TRANSCRIPTION FACTOR IIIA) |
| 1tf6_D | 2e-12 | 151 | 332 | 43 | 190 | PROTEIN (TRANSCRIPTION FACTOR IIIA) |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 554 | 562 | RPRGGRKRK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Essential protein (PubMed:22353599). Isoform 1 is a transcription activator the binds both 5S rDNA and 5S rRNA and stimulates the transcription of 5S rRNA gene (PubMed:12711688, PubMed:22353599). Isoform 1 regulates 5S rRNA levels during development (PubMed:22353599). {ECO:0000269|PubMed:12711688, ECO:0000269|PubMed:22353599}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00229 | DAP | Transfer from AT1G72050 | Download |
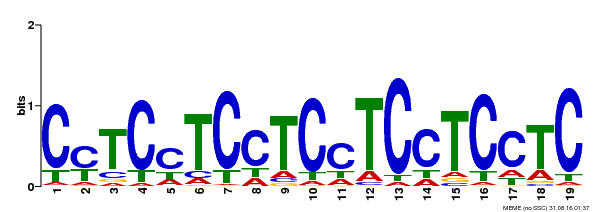
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 462846634 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025795605.1 | 0.0 | transcription factor IIIA-like isoform X1 | ||||
| Swissprot | Q84MZ4 | 1e-101 | TF3A_ARATH; Transcription factor IIIA | ||||
| TrEMBL | A0A2T7F0W9 | 0.0 | A0A2T7F0W9_9POAL; Uncharacterized protein | ||||
| TrEMBL | A0A2T8KTL1 | 0.0 | A0A2T8KTL1_9POAL; Uncharacterized protein | ||||
| STRING | Si020193m | 0.0 | (Setaria italica) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2382 | 38 | 82 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G72050.2 | 1e-103 | transcription factor IIIA | ||||



