 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 462854212 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Chloridoideae; Eragrostideae; Eragrostidinae; Eragrostis
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 189aa MW: 21439.3 Da PI: 10.853 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 51.7 | 2e-16 | 5 | 52 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rg+WT+eEd +l ++++G +W++ ++ g+ R++k+c++rw +yl
462854212 5 RGPWTPEEDRILTAHIERHGHSNWRALPKQAGLLRCGKSCRLRWINYL 52
89******************************99************97 PP
| |||||||
| 2 | Myb_DNA-binding | 53.9 | 4.2e-17 | 58 | 103 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rg++T eE++ +++++++lG++ W++Ia++++ gRt++++k+ w+++l
462854212 58 RGNFTREEEDAIIQLHHMLGNR-WSAIAARLP-GRTDNEIKNVWHTHL 103
89********************.*********.************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.26E-30 | 1 | 99 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 17.208 | 1 | 52 | IPR017930 | Myb domain |
| SMART | SM00717 | 2.5E-13 | 4 | 54 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.2E-14 | 5 | 52 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.0E-24 | 6 | 59 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 6.97E-11 | 7 | 52 | No hit | No description |
| PROSITE profile | PS51294 | 25.865 | 53 | 107 | IPR017930 | Myb domain |
| SMART | SM00717 | 3.2E-15 | 57 | 105 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 4.2E-16 | 58 | 103 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.66E-11 | 60 | 103 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 1.1E-26 | 60 | 107 | IPR009057 | Homeodomain-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 189 aa Download sequence Send to blast |
MGLKRGPWTP EEDRILTAHI ERHGHSNWRA LPKQAGLLRC GKSCRLRWIN YLRPDIKRGN 60 FTREEEDAII QLHHMLGNRW SAIAARLPGR TDNEIKNVWH THLKKRLEPK APTNAAPKRK 120 AKKQQQKQLQ PEVVVLDGPT TVTVSSPEQS LSTTTSAVTT DYSAASSMEN ADGFTSEEST 180 TASGRRRWR |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1h8a_C | 3e-26 | 3 | 107 | 25 | 128 | MYB TRANSFORMING PROTEIN |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Acts as negative regulator of cold tolerance. Negatively regulates beta-amylase genes at the transcriptional level in response to cold stress. Suppresses beta-amylase gene expression by interacting with TIFY11A/JAZ9. Maltose produced by beta-amylases has a role in protecting cell membranes under cold stress conditions in rice and may contribute to the cold tolerance as a compatible solute. {ECO:0000269|PubMed:28062835}. | |||||
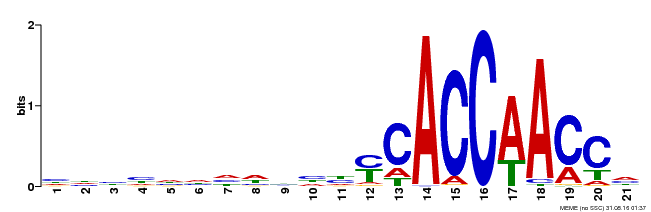
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00375 | DAP | Transfer from AT3G23250 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 462854212 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by cold stress and flooding. {ECO:0000269|PubMed:28062835}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | EU973471 | 1e-132 | EU973471.1 Zea mays clone 399373 myb-related protein Myb4 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025798785.1 | 1e-99 | transcription factor MYB30-like | ||||
| Swissprot | Q6K1S6 | 1e-81 | MYB30_ORYSJ; Transcription factor MYB30 | ||||
| TrEMBL | A0A2S3GRM6 | 3e-98 | A0A2S3GRM6_9POAL; Uncharacterized protein | ||||
| TrEMBL | A0A2T7F975 | 3e-98 | A0A2T7F975_9POAL; Uncharacterized protein | ||||
| STRING | GRMZM2G095904_P01 | 8e-95 | (Zea mays) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP79 | 38 | 563 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G23250.1 | 1e-72 | myb domain protein 15 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



