 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 462856678 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Chloridoideae; Eragrostideae; Eragrostidinae; Eragrostis
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 162aa MW: 17805.7 Da PI: 11.1275 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 46.6 | 7.4e-15 | 82 | 126 | 5 | 49 |
CHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 5 krerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkel 49
+r+rr++kNRe+A rsRqRK+a++ eLe ++++L++ N++L+k+
462856678 82 RRQRRMIKNRESAARSRQRKQAYMMELEAEIAKLKELNEELQKKQ 126
79*************************************997543 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00338 | 8.1E-12 | 78 | 143 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 10.955 | 80 | 125 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF00170 | 9.1E-13 | 82 | 128 | IPR004827 | Basic-leucine zipper domain |
| Gene3D | G3DSA:1.20.5.170 | 1.7E-14 | 82 | 135 | No hit | No description |
| SuperFamily | SSF57959 | 1.04E-10 | 82 | 128 | No hit | No description |
| CDD | cd14707 | 4.13E-23 | 82 | 134 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 85 | 100 | IPR004827 | Basic-leucine zipper domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009738 | Biological Process | abscisic acid-activated signaling pathway | ||||
| GO:0010255 | Biological Process | glucose mediated signaling pathway | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0000976 | Molecular Function | transcription regulatory region sequence-specific DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 162 aa Download sequence Send to blast |
MLFPQGNVFA PLVPPLSLGN GLVTGPVGQG GGGGAVSPVT SNGFGKMEGG DLSSLSPSPV 60 PFIFNGGLRG RKTSAIEKVA ERRQRRMIKN RESAARSRQR KQAYMMELEA EIAKLKELNE 120 ELQKKQVEML EMQKNEVLER MTRQIGPTAK RICLRRTLTG PW |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator that mediates abscisic acid (ABA) signaling (PubMed:18931143, PubMed:19947981, PubMed:27424498, PubMed:27325665). Can regulate the expression of a wide spectrum of stress-related genes in response to abiotic stresses through an ABA-dependent regulation pathway. Confers ABA-dependent drought and salinity tolerance (PubMed:18931143, PubMed:27325665). Binds specifically to the ABA-responsive elements (ABRE) in the promoter of target genes to mediate stress-responsive ABA signaling (PubMed:27325665, PubMed:27424498). {ECO:0000269|PubMed:18931143, ECO:0000269|PubMed:19947981, ECO:0000269|PubMed:27325665, ECO:0000269|PubMed:27424498}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00186 | DAP | Transfer from AT1G45249 | Download |
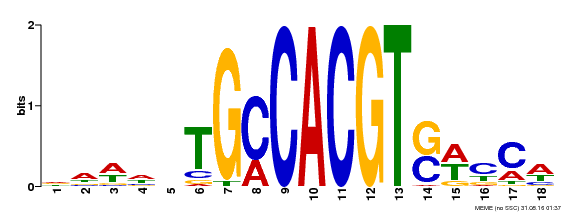
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 462856678 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by abscisic acid (ABA) (PubMed:18315698, PubMed:18931143). Induced by drought, salt and osmotic stresses (PubMed:18931143). {ECO:0000269|PubMed:18315698, ECO:0000269|PubMed:18931143}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004954029.1 | 3e-91 | bZIP transcription factor 23 | ||||
| Swissprot | Q6Z312 | 2e-85 | BZP23_ORYSJ; bZIP transcription factor 23 | ||||
| TrEMBL | K3YTP4 | 6e-90 | K3YTP4_SETIT; Uncharacterized protein | ||||
| STRING | OMERI02G31310.1 | 8e-85 | (Oryza meridionalis) | ||||
| STRING | LPERR02G27610.1 | 1e-85 | (Leersia perrieri) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP706 | 38 | 147 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G45249.1 | 2e-30 | abscisic acid responsive elements-binding factor 2 | ||||



