 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 462867517 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Chloridoideae; Eragrostideae; Eragrostidinae; Eragrostis
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 179aa MW: 19930.8 Da PI: 9.431 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 56.2 | 7.8e-18 | 13 | 60 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rg W++eEde+lv++++ +G +W+++++ g+ R++k+c++rw +yl
462867517 13 RGLWSPEEDEKLVRYITAHGHSCWSSVPKNAGLQRCGKSCRLRWINYL 60
788*******************************************97 PP
| |||||||
| 2 | Myb_DNA-binding | 48.3 | 2.4e-15 | 66 | 109 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqk 46
rg + +E+ +++d+++ lG++ W+ Ia++++ gRt++++k++w++
462867517 66 RGTLSDQEERIIIDVHRILGNK-WAQIAKHLP-GRTDNEVKNFWNS 109
678899****************.*********.***********96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 3.0E-26 | 4 | 63 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 17.488 | 8 | 60 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 6.68E-28 | 10 | 107 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 7.6E-13 | 12 | 62 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.7E-15 | 13 | 60 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.00E-10 | 16 | 60 | No hit | No description |
| PROSITE profile | PS51294 | 25.086 | 61 | 115 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 8.8E-24 | 64 | 116 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 7.8E-14 | 65 | 113 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.1E-13 | 66 | 109 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 3.76E-10 | 70 | 108 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 179 aa Download sequence Send to blast |
MGHHCCSKQK VKRGLWSPEE DEKLVRYITA HGHSCWSSVP KNAGLQRCGK SCRLRWINYL 60 RPDLKRGTLS DQEERIIIDV HRILGNKWAQ IAKHLPGRTD NEVKNFWNSC IKKKLIAQGL 120 DPKTHNLLPA SKSLLRGANP SSAHGVGANA AFDLELMESC GFFGGGAGGN AMDQLQWDC |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 1e-30 | 13 | 115 | 7 | 108 | B-MYB |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00350 | DAP | Transfer from AT3G12720 | Download |
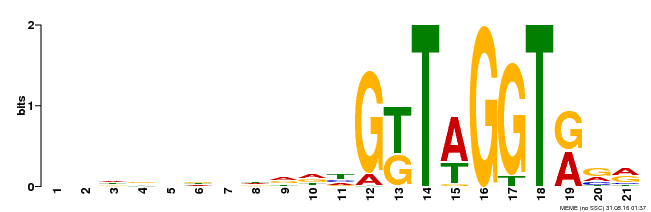
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 462867517 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AP004006 | 2e-52 | AP004006.5 Oryza sativa Japonica Group genomic DNA, chromosome 7, BAC clone:OJ1197_D06. | |||
| GenBank | AP004259 | 2e-52 | AP004259.2 Oryza sativa Japonica Group genomic DNA, chromosome 7, PAC clone:P0005E02. | |||
| GenBank | AP014963 | 2e-52 | AP014963.1 Oryza sativa Japonica Group DNA, chromosome 7, cultivar: Nipponbare, complete sequence. | |||
| GenBank | CP012615 | 2e-52 | CP012615.1 Oryza sativa Indica Group cultivar RP Bio-226 chromosome 7 sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004957753.1 | 2e-94 | transcription factor MYB61 | ||||
| Swissprot | Q8LPH6 | 3e-66 | MYB86_ARATH; Transcription factor MYB86 | ||||
| TrEMBL | K3ZU71 | 4e-93 | K3ZU71_SETIT; Uncharacterized protein | ||||
| STRING | Si030152m | 8e-94 | (Setaria italica) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP9751 | 35 | 45 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G12720.1 | 1e-84 | myb domain protein 67 | ||||



