 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 462884795 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Chloridoideae; Eragrostideae; Eragrostidinae; Eragrostis
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 238aa MW: 27372.5 Da PI: 10.1384 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 55.8 | 8.2e-18 | 48 | 94 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn ErrRRdriN+++ L+el+P++ K +Ka+iL +++eY+ksLq
462884795 48 VHNLSERRRRDRINEKLRALQELIPHC-----NKTDKASILDETIEYLKSLQ 94
6*************************8.....6******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF47459 | 2.49E-21 | 42 | 108 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 18.52 | 44 | 93 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 7.55E-16 | 47 | 98 | No hit | No description |
| Gene3D | G3DSA:4.10.280.10 | 1.3E-21 | 48 | 103 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 3.8E-15 | 48 | 94 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 2.0E-17 | 50 | 99 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009585 | Biological Process | red, far-red light phototransduction | ||||
| GO:0009693 | Biological Process | ethylene biosynthetic process | ||||
| GO:0010244 | Biological Process | response to low fluence blue light stimulus by blue low-fluence system | ||||
| GO:0010600 | Biological Process | regulation of auxin biosynthetic process | ||||
| GO:0010928 | Biological Process | regulation of auxin mediated signaling pathway | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 238 aa Download sequence Send to blast |
MSTRRRGGVG TLQIAGARNL QDAECETTEE TKSSRRYGSK RRTRAAEVHN LSERRRRDRI 60 NEKLRALQEL IPHCNKTDKA SILDETIEYL KSLQMQLQIM WMTSGMAPMM FPGAHQFMPP 120 MALGMNSACI PSAQSLGQMP RVPYMNHPLP NHFPLNSSPA MNSMNPPNVA NQMQNIHLRE 180 ISNHILHPDS DQTAAAQDPM LMDLKQYKRH NPVRYQNCQP VQLCRLLQRD NRLLMEFR |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 39 | 56 | KRRTRAAEVHNLSERRRR |
| 2 | 52 | 57 | ERRRRD |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that may act as negative regulator of phyB-dependent light signal transduction (PubMed:17485859). Transcription activator that acts as positive regulator of internode elongation. May function via regulation of cell wall-related genes. May play a role in a drought-associated growth-restriction mechanism in response to drought stress (PubMed:22984180). {ECO:0000269|PubMed:17485859, ECO:0000269|PubMed:22984180}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00082 | ChIP-seq | Transfer from AT3G59060 | Download |
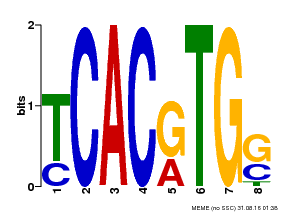
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 462884795 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by light in dark-grown etiolated seedlings (PubMed:17485859). Circadian oscillation under 12 h light/12 h dark cycle conditions, with peaks in the middle of the light period (PubMed:17485859, PubMed:22984180). Down-regulated by cold and drought stresses (PubMed:22984180). {ECO:0000269|PubMed:17485859, ECO:0000269|PubMed:22984180}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025793215.1 | 1e-105 | transcription factor PHYTOCHROME INTERACTING FACTOR-LIKE 13-like | ||||
| Refseq | XP_025793216.1 | 1e-105 | transcription factor PHYTOCHROME INTERACTING FACTOR-LIKE 13-like | ||||
| Refseq | XP_025793217.1 | 1e-105 | transcription factor PHYTOCHROME INTERACTING FACTOR-LIKE 13-like | ||||
| Swissprot | Q10CH5 | 1e-100 | PIL13_ORYSJ; Transcription factor PHYTOCHROME INTERACTING FACTOR-LIKE 13 | ||||
| TrEMBL | A0A1E5URH3 | 1e-106 | A0A1E5URH3_9POAL; Transcription factor PIF5 | ||||
| STRING | Si035818m | 1e-104 | (Setaria italica) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP3174 | 34 | 82 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G59060.4 | 3e-38 | phytochrome interacting factor 3-like 6 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



