 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 462900609 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Chloridoideae; Eragrostideae; Eragrostidinae; Eragrostis
|
||||||||
| Family | GATA | ||||||||
| Protein Properties | Length: 158aa MW: 17396.9 Da PI: 9.9308 | ||||||||
| Description | GATA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | GATA | 63.8 | 2e-20 | 28 | 62 | 1 | 35 |
GATA 1 CsnCgttkTplWRrgpdgnktLCnaCGlyyrkkgl 35
C+ C+ttkTplWR gp+g+k+LCnaCG++yrk+++
462900609 28 CADCHTTKTPLWRGGPEGPKSLCNACGIRYRKRRR 62
********************************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF57716 | 9.98E-14 | 20 | 61 | No hit | No description |
| PROSITE profile | PS50114 | 13.231 | 22 | 58 | IPR000679 | Zinc finger, GATA-type |
| SMART | SM00401 | 2.5E-18 | 22 | 79 | IPR000679 | Zinc finger, GATA-type |
| CDD | cd00202 | 2.53E-15 | 27 | 63 | No hit | No description |
| Gene3D | G3DSA:3.30.50.10 | 2.6E-16 | 27 | 62 | IPR013088 | Zinc finger, NHR/GATA-type |
| PROSITE pattern | PS00344 | 0 | 28 | 53 | IPR000679 | Zinc finger, GATA-type |
| Pfam | PF00320 | 3.1E-18 | 28 | 62 | IPR000679 | Zinc finger, GATA-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 158 aa Download sequence Send to blast |
MEAEMETDPI SPSPDDTTTA SGGEAKACAD CHTTKTPLWR GGPEGPKSLC NACGIRYRKR 60 RRQALGLDAN AEPQLDQQQK KKAAAAAASS SKEDKKKEDK KEEGDKKKQV TVELRVVGFG 120 KEVMLKQRRR MRRKKCLSEE ERAAVLLMSL SSGVIYAS |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 57 | 61 | RKRRR |
| 2 | 126 | 132 | QRRRMRR |
| 3 | 127 | 134 | RRRMRRKK |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00335 | DAP | Transfer from AT3G06740 | Download |
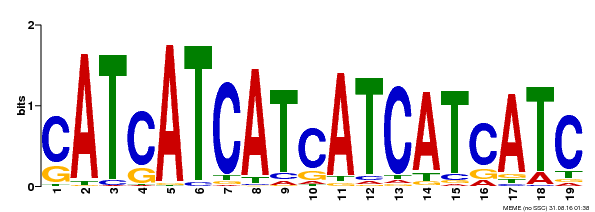
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 462900609 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | FP098913 | 2e-36 | FP098913.1 Phyllostachys edulis cDNA clone: bphyst028n16, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025818098.1 | 9e-64 | GATA transcription factor 15-like | ||||
| TrEMBL | A0A0A9NUC3 | 2e-79 | A0A0A9NUC3_ARUDO; Uncharacterized protein | ||||
| STRING | Pavir.Eb04066.1.p | 2e-61 | (Panicum virgatum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2418 | 38 | 92 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G06740.1 | 7e-21 | GATA transcription factor 15 | ||||



