 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 462917549 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Chloridoideae; Eragrostideae; Eragrostidinae; Eragrostis
|
||||||||
| Family | ARF | ||||||||
| Protein Properties | Length: 705aa MW: 77692.6 Da PI: 8.2235 | ||||||||
| Description | ARF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 76.9 | 2.2e-24 | 134 | 235 | 1 | 99 |
EEEE-..-HHHHTT-EE--HHH.HTT......---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SSSEE..EE CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh......ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldgrsefelvv 94
f+k+lt sd++++g +++p++ ae++ +++ +++ +l+ +d++g +W++++iyr++++r++lt+GW+ Fv+ ++L +gD v+F + +++el++
462917549 134 FCKTLTASDTSTHGGFSVPRRAAEDCfppldySQQRPSP-ELVAKDLHGTEWKFRHIYRGQPRRHLLTTGWSAFVNKKKLVSGDAVLFL--RGDDGELRL 230
99****************************975454455.9************************************************..3489999** PP
EEE-S CS
B3 95 kvfrk 99
+v+r+
462917549 231 GVRRA 235
*9996 PP
| |||||||
| 2 | Auxin_resp | 111.7 | 7.3e-37 | 260 | 340 | 1 | 82 |
Auxin_resp 1 aahaastksvFevvYnPrastseFvvkvekvekalkvkvsvGmRfkmafetedsserrlsGtvvgvsdldpvrWpnSkWrsL 82
+aha++tks F+++YnPr s+ eF+++++k++++l++++s+GmRfkm++e+ed+ err++G+++g+s++dp W+ SkW++L
462917549 260 VAHAVATKSEFHIYYNPRLSHCEFIIPYSKFMRSLSQPFSAGMRFKMRYESEDATERRYTGIITGISEADP-TWRASKWKCL 340
79*********************************************************************.9********9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101936 | 1.7E-42 | 128 | 250 | IPR015300 | DNA-binding pseudobarrel domain |
| Gene3D | G3DSA:2.40.330.10 | 1.7E-39 | 129 | 237 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 7.63E-22 | 132 | 234 | No hit | No description |
| PROSITE profile | PS50863 | 13.31 | 134 | 236 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02362 | 4.4E-22 | 134 | 235 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 2.5E-23 | 134 | 236 | IPR003340 | B3 DNA binding domain |
| Pfam | PF06507 | 1.8E-30 | 260 | 340 | IPR010525 | Auxin response factor |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009725 | Biological Process | response to hormone | ||||
| GO:0010050 | Biological Process | vegetative phase change | ||||
| GO:0010158 | Biological Process | abaxial cell fate specification | ||||
| GO:0010582 | Biological Process | floral meristem determinacy | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 705 aa Download sequence Send to blast |
MRRRRRRPPT LAEDPIXXXX PPTPQPRPGA AVCLELWHAC AGPVAPLPRK GSVVVYLPQG 60 HLEQLGDAAS GGGGADVTLH ADAATDEVYA QLALVDENEE MARRLRGGSE DGSGGDADDG 120 DTVKQRFARM PHMFCKTLTA SDTSTHGGFS VPRRAAEDCF PPLDYSQQRP SPELVAKDLH 180 GTEWKFRHIY RGQPRRHLLT TGWSAFVNKK KLVSGDAVLF LRGDDGELRL GVRRAAQLKN 240 GSAFPALYNQ CSNLGTLANV AHAVATKSEF HIYYNPRLSH CEFIIPYSKF MRSLSQPFSA 300 GMRFKMRYES EDATERRYTG IITGISEADP TWRASKWKCL MVRWDDDVDF RRSNRVSPWE 360 VELTGSVTGS HLSTPNSKRL KPCLSHANAD LLVPHGSGCP DFAESAQFHK VLQGQELHGY 420 RTHNSTAAAA QHCEARNMQY IDERSCSSDV SNSIPGVPRL GARAPLGSPG FSYHCSGFGE 480 SQRFQKVLQG QEVLRPYRGP QVDACMRTGS FHQQDGPLAP SVANKWHAQL NGCVYRGPPA 540 SLLPSQTSSP PSVLMFQQSL SKMSQFEFGN AYMDQNGDDR SAMFGHVEGI GRTEQMLMTQ 600 PHNVYGEVAN GHLTVERLHS AIGIGKDVPD NREVRTNSCK LFGISLTEKV PARKEMDCGD 660 ASYPSPFQSL KQQVPKSLGN SCATVHEQRP VVGRVIDVST MDMMI |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4ldv_A | 1e-105 | 31 | 370 | 18 | 361 | Auxin response factor 1 |
| 4ldw_A | 1e-105 | 31 | 370 | 18 | 361 | Auxin response factor 1 |
| 4ldw_B | 1e-105 | 31 | 370 | 18 | 361 | Auxin response factor 1 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 1 | 6 | RRRRRR |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Auxin response factors (ARFs) are transcriptional factors that bind specifically to the DNA sequence 5'-TGTCTC-3' found in the auxin-responsive promoter elements (AuxREs). | |||||
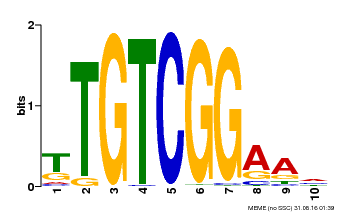
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00021 | PBM | Transfer from AT2G33860 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 462917549 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025807170.1 | 0.0 | auxin response factor 15-like | ||||
| Swissprot | Q8S985 | 0.0 | ARFO_ORYSJ; Auxin response factor 15 | ||||
| TrEMBL | A0A1E5W003 | 0.0 | A0A1E5W003_9POAL; Auxin response factor | ||||
| STRING | Pavir.J08401.1.p | 0.0 | (Panicum virgatum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2537 | 35 | 80 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G33860.1 | 1e-155 | ARF family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



