 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 462920610 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Chloridoideae; Eragrostideae; Eragrostidinae; Eragrostis
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 526aa MW: 56719.9 Da PI: 4.8657 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 54.5 | 2.6e-17 | 16 | 63 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT Ed +lvd+vk++G g+W+++ + g+ R++k+c++rw ++l
462920610 16 KGPWTSAEDAILVDYVKKHGEGNWNAVQKNTGLSRCGKSCRLRWANHL 63
79******************************************9986 PP
| |||||||
| 2 | Myb_DNA-binding | 52.8 | 9.4e-17 | 69 | 112 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqk 46
+g++T+eE+ l++++++++G++ W++ a++++ gRt++++k++w++
462920610 69 KGAFTPEEERLIIQLHAKMGNK-WARMAAHLP-GRTDNEIKNYWNT 112
799*******************.*********.***********96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 18.19 | 11 | 63 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.16E-31 | 13 | 110 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 6.1E-15 | 15 | 65 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 2.8E-15 | 16 | 63 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.7E-24 | 17 | 70 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 2.02E-11 | 18 | 63 | No hit | No description |
| PROSITE profile | PS51294 | 25.454 | 64 | 118 | IPR017930 | Myb domain |
| SMART | SM00717 | 1.7E-17 | 68 | 116 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 5.7E-16 | 69 | 112 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 3.0E-26 | 71 | 116 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 6.41E-13 | 71 | 112 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009789 | Biological Process | positive regulation of abscisic acid-activated signaling pathway | ||||
| GO:0043068 | Biological Process | positive regulation of programmed cell death | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0045926 | Biological Process | negative regulation of growth | ||||
| GO:0048235 | Biological Process | pollen sperm cell differentiation | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 526 aa Download sequence Send to blast |
PADDGGSPNR GPSLKKGPWT SAEDAILVDY VKKHGEGNWN AVQKNTGLSR CGKSCRLRWA 60 NHLRPNLKKG AFTPEEERLI IQLHAKMGNK WARMAAHLPG RTDNEIKNYW NTRVKRCQRA 120 GLPIYPASVC NQSSNEDEQV SDDFNCGDNL GSGLLNGNGL FLPDFTSDNF IGNPDALTYA 180 PQLSAVSISN LLGQSFASRS CSFLDHVDQT GILKHSGSVL PSLSDTVDGV LSSVDHFSND 240 SEKLKQALGF DYLSEATACS KTIAPFGVAL SGSHAFLNGT FSASRPTNGP LKMELPSLQD 300 TESDPISWLK YTVAPAMQPT ELVDPYLQSP TATPSVKSEC ASPRNSGLLE ELLHEAQVLR 360 SGKNQQLSVR SSSSSADTPC ENATAVSAEF DICQEYWEEH PSSFPIEYTP FSGNSFTEST 420 PPVSAASPDI FQFSKISPAQ SPSMGSGDPV TEPKYESVGS PHPDNFRPDA LFSGNTTDVS 480 IFNDAIAMLL GNGVSADSKP VLGDGTAFNY STWSNMPHVC EMSEFK |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1h8a_C | 4e-33 | 12 | 116 | 23 | 126 | MYB TRANSFORMING PROTEIN |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator of gibberellin-dependent alpha-amylase expression in aleurone cells. Involved in pollen and floral organs development. May bind to the 5'-TAACAAA-3' box of alpha-amylase promoter. {ECO:0000269|PubMed:9150608}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00490 | DAP | Transfer from AT5G06100 | Download |
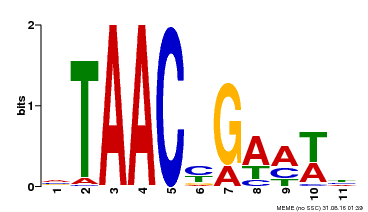
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 462920610 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By gibberellin in aleurone cells. {ECO:0000269|PubMed:9150608}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025819835.1 | 0.0 | transcription factor GAMYB-like | ||||
| Swissprot | A2WW87 | 0.0 | GAM1_ORYSI; Transcription factor GAMYB | ||||
| TrEMBL | A0A2S3HQQ9 | 0.0 | A0A2S3HQQ9_9POAL; Transcription factor | ||||
| TrEMBL | A0A2T7DFC7 | 0.0 | A0A2T7DFC7_9POAL; Transcription factor | ||||
| STRING | Pavir.Ea03206.1.p | 0.0 | (Panicum virgatum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP3947 | 36 | 64 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G11440.1 | 2e-85 | myb domain protein 65 | ||||



