 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 462924164 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Chloridoideae; Eragrostideae; Eragrostidinae; Eragrostis
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 271aa MW: 29324.7 Da PI: 6.7505 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 51.5 | 1.8e-16 | 108 | 154 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn E+rRR+riN+++ L++l+P++ K +Ka++L +A+eY+k+Lq
462924164 108 VHNLSEKRRRSRINEKMKALQSLIPNS-----NKTDKASMLDEAIEYLKQLQ 154
6*************************7.....5******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00083 | 1.27E-18 | 100 | 158 | No hit | No description |
| SuperFamily | SSF47459 | 9.81E-21 | 101 | 164 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 7.0E-20 | 101 | 164 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 18.063 | 104 | 153 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 6.5E-14 | 108 | 154 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 4.0E-18 | 110 | 159 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0007623 | Biological Process | circadian rhythm | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0010114 | Biological Process | response to red light | ||||
| GO:0010154 | Biological Process | fruit development | ||||
| GO:0010187 | Biological Process | negative regulation of seed germination | ||||
| GO:0048440 | Biological Process | carpel development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 271 aa Download sequence Send to blast |
MHEQGLGGFG GSRAAVQGHG GRDAMLFLQQ HHHQQQQRQQ QQLEEAEEEE ARRQMFAGVA 60 FPGALGYGHQ AEEAGGLGDS DAGGSEPEPA PSRARGGSGS KRSRAAEVHN LSEKRRRSRI 120 NEKMKALQSL IPNSNKTDKA SMLDEAIEYL KQLQLQVQML SMRNGVYLNP PYLSGALEAA 180 HASQMFAALG GGNITGPSSG AAMLPVNQSS GAQHQAFEPL NPPPQNQQAT LILPSVPDKT 240 IPEPPFHLES SQSHLRSFQM PESSEVIRLR E |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00035 | PBM | Transfer from AT4G36930 | Download |
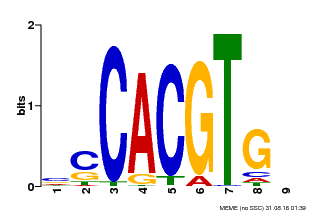
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 462924164 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT035360 | 1e-117 | BT035360.1 Zea mays full-length cDNA clone ZM_BFb0057H06 mRNA, complete cds. | |||
| GenBank | KJ726860 | 1e-117 | KJ726860.1 Zea mays clone pUT3401 bHLH transcription factor (bHLH125) mRNA, partial cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025811540.1 | 1e-116 | uncharacterized protein LOC112889235 isoform X1 | ||||
| TrEMBL | A0A2T7E2L4 | 1e-115 | A0A2T7E2L4_9POAL; Uncharacterized protein | ||||
| STRING | Pavir.J38250.1.p | 1e-111 | (Panicum virgatum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP3336 | 37 | 76 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G36930.1 | 6e-35 | bHLH family protein | ||||



