 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 462927945 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Chloridoideae; Eragrostideae; Eragrostidinae; Eragrostis
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 800aa MW: 86727.2 Da PI: 8.7189 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 38.9 | 1.6e-12 | 650 | 696 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCC.C...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPk.askapskKlsKaeiLekAveYIksLq 55
+h e+Er+RR+++N++f Lr ++P+ + K++Ka+ L A+ YI +Lq
462927945 650 NHVEAERQRREKLNQRFYALRAVVPNiS------KMDKASLLGDAITYITDLQ 696
799***********************66......*****************99 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF14215 | 7.1E-50 | 245 | 430 | IPR025610 | Transcription factor MYC/MYB N-terminal |
| PROSITE profile | PS50888 | 16.831 | 646 | 695 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 5.63E-19 | 649 | 714 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 7.54E-15 | 649 | 700 | No hit | No description |
| Gene3D | G3DSA:4.10.280.10 | 4.1E-18 | 650 | 709 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 5.2E-10 | 650 | 696 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.9E-16 | 652 | 701 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 800 aa Download sequence Send to blast |
MTHVPQAHSV PAREPSRGED GSQLGWDPSS AVSSKARKGS SRKEEEGVVV LPPSLPSIHP 60 GSTGGACTLG SRNPLPHQRN SATTSISFLN ERPASTHEVL SLLFAPVLGF TREEIRAPRL 120 GFHPPARAGF RGLGVDSGAG PSIRCTGDLF FFTPRVRREL IRPSWNLIGP LLFGVIDYIL 180 VHSIMVMKME IEDDGAAGGG TGGTWTEEDR ALGAAVLGTD AFAYLTKGGG AISQGLVATS 240 LSVDMQNRLQ ELVESDRPGA GWNYAIYWQL SRTKSGDLVL GWGDGSCREP RDGEVAAAAS 300 EGSDDTKQRM RKRVLQRLHI AFGGADEEDY ALGIDQVTDT EVFFLASMYF AFPRRVGGPG 360 QVFAAGIPLW IPNSERKVYP TNYCYRGFLA NAAGFRTIVL VPFESGVLEL GSMQHIAESP 420 DVIETIRSVF AGASGNKAAV QKPEGNGSTP TERSPSLAKI FGKDLNLGRP SAAPVVSVSK 480 VDERSWEQRS AAGGSSLLPN VPKVSPSYNW SQARGLNSHQ QKFGNGVLIV SNEAAQRSNG 540 AADSSSATHF QLQKVPPLQK LQLQKLPQIQ KPPPLVTQQQ LQPQVPRQID FSAGSSSKPG 600 VLVTRTSVLD GDSAEVDGLC KEEGPPPIVE DRRPRKRGRK PANGREEPLN HVEAERQRRE 660 KLNQRFYALR AVVPNISKMD KASLLGDAIT YITDLQKKLK EMETERERLL ESGMMGGTPR 720 PEVDIQVVQD EVLVRVMSPM ENHPVKKVFQ AFEEAEVRIG ESKVTGNNGT VVHSFIIKCP 780 GTEQQTREKV IAAMSRAMSS |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5gnj_A | 6e-27 | 644 | 706 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_B | 6e-27 | 644 | 706 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_E | 6e-27 | 644 | 706 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_F | 6e-27 | 644 | 706 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_G | 6e-27 | 644 | 706 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_I | 6e-27 | 644 | 706 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_M | 6e-27 | 644 | 706 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_N | 6e-27 | 644 | 706 | 2 | 64 | Transcription factor MYC2 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 631 | 639 | RRPRKRGRK |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00100 | PBM | Transfer from AT1G01260 | Download |
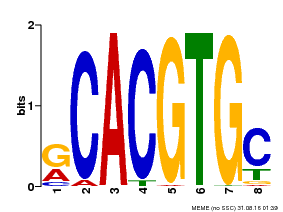
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 462927945 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025816895.1 | 0.0 | transcription factor bHLH13-like | ||||
| TrEMBL | A0A3L6SNM9 | 0.0 | A0A3L6SNM9_PANMI; Transcription factor bHLH13-like | ||||
| STRING | Pavir.Ea00030.1.p | 0.0 | (Panicum virgatum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP5174 | 38 | 60 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G01260.3 | 5e-67 | bHLH family protein | ||||



