 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 462931397 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Chloridoideae; Eragrostideae; Eragrostidinae; Eragrostis
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 701aa MW: 78531.8 Da PI: 6.7277 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 113.9 | 9.1e-36 | 2 | 77 | 1 | 76 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkk 76
+Cqv+gCead+ e+k yhrrh+vC +++a++v+++g+++r+CqqC++fh l +fDe+krsCrr+L++hn+rrr+k
462931397 2 RCQVPGCEADIRELKGYHRRHRVCLLCAHAAAVMLDGVQKRYCQQCGKFHILLDFDEDKRSCRRKLERHNKRRRRK 77
6************************************************************************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51141 | 29.624 | 1 | 77 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 1.44E-33 | 1 | 79 | IPR004333 | Transcription factor, SBP-box |
| Gene3D | G3DSA:4.10.1100.10 | 3.4E-26 | 2 | 64 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 4.3E-27 | 3 | 77 | IPR004333 | Transcription factor, SBP-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0055070 | Biological Process | copper ion homeostasis | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 701 aa Download sequence Send to blast |
MRCQVPGCEA DIRELKGYHR RHRVCLLCAH AAAVMLDGVQ KRYCQQCGKF HILLDFDEDK 60 RSCRRKLERH NKRRRRKPGS NGPFEKEMDE HFDLSADISG DGELREGNLP IENIEGTTSE 120 MLETVLSNKV LDRETPVGSE DVLSSPTCTE PTLQNEQSKS MVTFAASVEA CIGAKQENMK 180 LNSSPVHDTK SAYSSSCPTG RISFKLYDWN PAEFPRRLRH QIFEWLASMP VELEGYIRPG 240 CTILTVFIAM PQHMWDKLSD DTANLLRNLV NSPTSLLLGK GAFFIHTDNM IFQVLKDGAT 300 LMSSKIEVQA PRIHYVHPTW FEAGKPVELH LCGSSLDQPK LRSLVSFDGE YLKHDCFRLS 360 SLDAFDCVEK GDLTLDSQHE IFRICIKESR SDTHGPAFVE VENMFGLSNF VPILFGSKQL 420 CSELEKIQAV LCGSCKNNKV LCGSSCKNSD IVGEFPDASF DPCERQKIQS AAMSEFLVDI 480 GWLIRKPTPE EFKNLLSLTN IQRWVNMLKF LIQNDFFNVL EIIVKSVDNI VGSEIISNLE 540 RGRLEDHVTT FLGYVDHARS IIHHRAKHGE GTQLESRLVT DNFLKQPSLG TSVPLDNGNT 600 GPCGDNYLQS TSAACEEEVT VPLMTKDASH RQCCHPEINA RWLKPSLVIT FPGGATRTRL 660 LTSVVVAAVL CFTACVVLFH SHRVGMLAAP VKRYLSSDSA S |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul5_A | 3e-36 | 2 | 82 | 5 | 85 | squamosa promoter binding protein-like 7 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 59 | 77 | KRSCRRKLERHNKRRRRKP |
| 2 | 63 | 75 | RRKLERHNKRRRR |
| 3 | 70 | 77 | NKRRRRKP |
| 4 | 71 | 76 | KRRRRK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3'. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00604 | PBM | Transfer from AT5G18830 | Download |
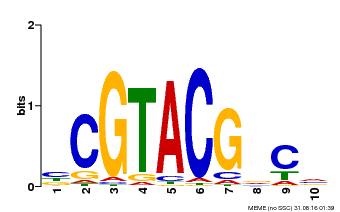
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 462931397 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025805456.1 | 0.0 | squamosa promoter-binding-like protein 9 | ||||
| Swissprot | Q6I576 | 0.0 | SPL9_ORYSJ; Squamosa promoter-binding-like protein 9 | ||||
| TrEMBL | A0A2T8KJS3 | 0.0 | A0A2T8KJS3_9POAL; Uncharacterized protein | ||||
| STRING | Sb09g020110.1 | 0.0 | (Sorghum bicolor) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP6922 | 33 | 44 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G18830.3 | 1e-134 | squamosa promoter binding protein-like 7 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



