 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 462940689 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Chloridoideae; Eragrostideae; Eragrostidinae; Eragrostis
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 427aa MW: 47157.8 Da PI: 6.8401 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 25.6 | 2.9e-08 | 24 | 50 | 1 | 28 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIa 28
r rWT+ E++++++a k++G W +I
462940689 24 RERWTEAEHKRFLEALKLYGRQ-WQRIE 50
78******************77.**996 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 8.97E-7 | 18 | 63 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.1E-7 | 22 | 51 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 0.0076 | 23 | 75 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.0E-4 | 24 | 50 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 0.00525 | 26 | 64 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009739 | Biological Process | response to gibberellin | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0042754 | Biological Process | negative regulation of circadian rhythm | ||||
| GO:0043433 | Biological Process | negative regulation of sequence-specific DNA binding transcription factor activity | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0048574 | Biological Process | long-day photoperiodism, flowering | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 427 aa Download sequence Send to blast |
MEVNSSGEET MIKVRKPYTI TKQRERWTEA EHKRFLEALK LYGRQWQRIE VCIASSSWSP 60 LQIYGRERIK LQLEKEAMNN GTSPGQAHDI DIPPPRPKRK PNCPYPRKSG LSSEISNKEI 120 PNDKSTKSNM RPSNSSVEMA SDSSLQKLQR KEVSEKGSCS EVLNLFRDVP SASFSSVNKS 180 SSNHGAARGT EPTKNEIKGM ATMENNSLSI NMHEDEKQIN DQEMERLNGI HISSKCDRFD 240 EDYLDSSTQQ MKLKPMSMEA ANVDRQTSRA SHCVTERNEA VSIPVMGTEV SHPDQISDQE 300 GANGSMNPCI HPTLSVDAKF NINATPLPCP HNFAAFPPLM QCNCNQDAYR SFVNMSSTFS 360 SMLVSTLLSN PAIHAAARLA ASYWPAAESN TSIDPNQDNP AEGVQGSDIG SPPSMASMVA 420 ATVGSRK |
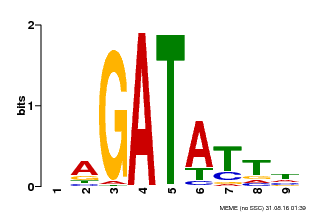
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00654 | PBM | Transfer from LOC_Os08g06110 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 462940689 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025820794.1 | 0.0 | protein LHY isoform X1 | ||||
| Refseq | XP_025820795.1 | 0.0 | protein LHY isoform X1 | ||||
| TrEMBL | A0A2T8IF63 | 0.0 | A0A2T8IF63_9POAL; Uncharacterized protein | ||||
| STRING | Pavir.Fa01905.1.p | 0.0 | (Panicum virgatum) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G46830.1 | 4e-26 | circadian clock associated 1 | ||||



