 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 462950116 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Chloridoideae; Eragrostideae; Eragrostidinae; Eragrostis
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 637aa MW: 69779.1 Da PI: 6.872 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 105.5 | 2.8e-33 | 153 | 209 | 2 | 59 |
--SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
dDgynWrKYGqK+vkgse+prsYY+Ct+++Cp+kkkvers+ d++++ei+Y+g+Hnh
462950116 153 DDGYNWRKYGQKQVKGSENPRSYYKCTFPSCPTKKKVERSQ-DGQITEIVYKGTHNHA 209
8****************************************.***************7 PP
| |||||||
| 2 | WRKY | 105.8 | 2.3e-33 | 308 | 366 | 1 | 59 |
---SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 1 ldDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
ldDgy+WrKYGqK+vkg+++prsYY+Ct+a+Cpv+k+ver+++d ++v++tYeg+Hnh+
462950116 308 LDDGYRWRKYGQKVVKGNPNPRSYYKCTTANCPVRKHVERASHDLRAVITTYEGKHNHD 366
59********************************************************7 PP
| |||||||
| 3 | WRKY | 105.8 | 2.3e-33 | 480 | 538 | 1 | 59 |
---SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 1 ldDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
ldDgy+WrKYGqK+vkg+++prsYY+Ct+a+Cpv+k+ver+++d ++v++tYeg+Hnh+
462950116 480 LDDGYRWRKYGQKVVKGNPNPRSYYKCTTANCPVRKHVERASHDLRAVITTYEGKHNHD 538
59********************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 2.2E-28 | 144 | 211 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 23.755 | 147 | 211 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 4.58E-25 | 148 | 211 | IPR003657 | WRKY domain |
| SMART | SM00774 | 1.1E-35 | 152 | 210 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 2.3E-25 | 153 | 208 | IPR003657 | WRKY domain |
| Gene3D | G3DSA:2.20.25.80 | 5.3E-37 | 293 | 368 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 2.22E-29 | 300 | 368 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 37.883 | 303 | 368 | IPR003657 | WRKY domain |
| SMART | SM00774 | 2.0E-38 | 308 | 367 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 1.6E-25 | 309 | 366 | IPR003657 | WRKY domain |
| Gene3D | G3DSA:2.20.25.80 | 5.3E-37 | 465 | 540 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 2.22E-29 | 472 | 540 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 37.883 | 475 | 540 | IPR003657 | WRKY domain |
| SMART | SM00774 | 2.0E-38 | 480 | 539 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 1.6E-25 | 481 | 538 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0010120 | Biological Process | camalexin biosynthetic process | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0010508 | Biological Process | positive regulation of autophagy | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:0070370 | Biological Process | cellular heat acclimation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 637 aa Download sequence Send to blast |
SFFNIPGGLN PADFLDSPIL LTSSLFPSPT AGAFASQQFS WMQTQAVEQD GSKEEQRQQS 60 FPDFSFQTAP TSQEAVRTTT TTFQPPIQPA SLGEEAYTSQ QQQPWGYQQQ QPAMDTNQAS 120 FSAPFQATSS DATAMAPHAA QGYSQQSQRR SSDDGYNWRK YGQKQVKGSE NPRSYYKCTF 180 PSCPTKKKVE RSQDGQITEI VYKGTHNHAK PQNTRRNNSG AAQLLVQGGG DASEHSFGGT 240 PVGTPENSSA SFGEDEAGNA GGEEFDEDEP DSKRWRKESE GEGMSMAGNR TVREPRVVVQ 300 TMSDIDILDD GYRWRKYGQK VVKGNPNPRS YYKCTTANCP VRKHVERASH DLRAVITTYE 360 GKHNHDVPAA RGSAALYRPA PPPPPADNGA GSSGHHYLGG GGDASEHSFG GTPVGTPENS 420 SASFGEDEAG NAGGEEFDED EPDSKRWRKE SEGEGMSMAG NRTVREPRVV VQTMSDIDIL 480 DDGYRWRKYG QKVVKGNPNP RSYYKCTTAN CPVRKHVERA SHDLRAVITT YEGKHNHDVP 540 AARGSAALYR PAPPPPPADN GAGSSGQQRR RRAAVRPRSS FGLQQGAAGF GGFSGFDNPM 600 GSYMSQHQQQ QRQNDAMHAS RAKEEPREDM FFQQTMM |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj2_A | 9e-35 | 140 | 541 | 5 | 78 | Probable WRKY transcription factor 4 |
| 2lex_A | 9e-35 | 140 | 541 | 5 | 78 | Probable WRKY transcription factor 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator (PubMed:26025535). Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element (PubMed:19199048, PubMed:26025535). Negative regulator of both gibberellic acid (GA) and abscisic acid (ABA) signaling in aleurone cells, probably by interfering with GAM1, via the specific repression of GA- and ABA-induced promoters (PubMed:15618416, PubMed:19199048, PubMed:26025535). {ECO:0000269|PubMed:15618416, ECO:0000269|PubMed:19199048, ECO:0000269|PubMed:26025535}. | |||||
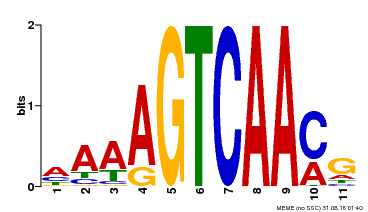
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00299 | DAP | Transfer from AT2G38470 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 462950116 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by abscisic acid (ABA) in aleurone cells, embryos, roots and leaves (PubMed:15618416, PubMed:19199048). Slightly down-regulated by gibberellic acid (GA) (PubMed:15618416). Accumulates in response to jasmonic acid (MeJA) (By similarity). {ECO:0000250|UniProtKB:Q6B6R4, ECO:0000269|PubMed:15618416, ECO:0000269|PubMed:19199048}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK357671 | 1e-175 | AK357671.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv1058P10. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004970421.3 | 0.0 | WRKY transcription factor WRKY24 | ||||
| Swissprot | Q6IEQ7 | 0.0 | WRK24_ORYSJ; WRKY transcription factor WRKY24 | ||||
| TrEMBL | A0A1E5WCJ0 | 0.0 | A0A1E5WCJ0_9POAL; Putative WRKY transcription factor 33 | ||||
| STRING | Pavir.Eb03590.1.p | 0.0 | (Panicum virgatum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2992 | 36 | 87 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G38470.1 | 1e-83 | WRKY DNA-binding protein 33 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



