 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 462951446 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Chloridoideae; Eragrostideae; Eragrostidinae; Eragrostis
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 972aa MW: 106079 Da PI: 5.92 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 129.8 | 9.6e-41 | 151 | 228 | 1 | 78 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
+CqvegC adl+ ak+yhrrhkvCe+h+ka++++v ++ qrfCqqCsrfh l+efDe krsCrrrLa+hn+rrrk+++
462951446 151 ACQVEGCGADLTAAKDYHRRHKVCEMHAKASTAVVGNTVQRFCQQCSRFHLLQEFDEWKRSCRRRLAGHNRRRRKTRP 228
5**************************************************************************875 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 1.2E-33 | 144 | 213 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 32.291 | 149 | 226 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 1.7E-38 | 150 | 230 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 9.7E-30 | 152 | 225 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS50297 | 10.02 | 756 | 823 | IPR020683 | Ankyrin repeat-containing domain |
| SMART | SM00248 | 0.15 | 756 | 785 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF48403 | 2.02E-8 | 759 | 863 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 4.4E-8 | 760 | 864 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 3.02E-10 | 761 | 861 | No hit | No description |
| SMART | SM00248 | 410 | 802 | 832 | IPR002110 | Ankyrin repeat |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 972 aa Download sequence Send to blast |
MEAARLGAQS SHLYGGGLVE LDMNRREKRV FGWDLNDWSW DSERFVATPV PSSVANGSAL 60 NSSPSSSEEA EAEAARNGAV RGESDKRKRV VVIDDDGTED QDLMGNSSGA LSLRIGGDSV 120 GAGTMENDGV NEEERNGKKI KVQGGSSSGP ACQVEGCGAD LTAAKDYHRR HKVCEMHAKA 180 STAVVGNTVQ RFCQQCSRFH LLQEFDEWKR SCRRRLAGHN RRRRKTRPDI GLGGAASIED 240 KVSNYLLLSL LGICANLNSG NSEHSSGQEL LSNLLRNLGT VAKSLEPKEL SKLLEACQSL 300 QNGSNAGISG TANALVNNSA AEAAGPSNSK PLFMNGNQCG QTSSSAMPIQ SKAAMAAIPE 360 PRACKVKDFD LNDTCDDMEG FENGEEGSPT PAFMTADSPN CPSWMQQDST QSPPQTSGNS 420 DSTSTQSLSS SNGDAQCRTD KIVFKLFEKV PSDLPPVLRS QILGWLSSSP TDIESYIRPG 480 CVILTVYLRL VESSWRELSD NMSSHLDKLL NSSSDNFWAS GLVLVMVRHQ LAFMYNGRVM 540 LDRPLAPSSY HYCRILCIKP VAAPYSAAVN FKVEGINLIN ASSRVICSFE GRCLFQEDTA 600 IVADDAELED RDTECLSFCC PLPGSRGRGF IEVEDSGFSN GFFPFIIAEQ DVCSEVTELE 660 SIFESSSHEL ADVDDVARSQ ALEFLNELGW LLHRANSTSK HDKTEPSLAT FNLWRFRNLG 720 IFAMEREWCA VMKMLLDFLF IGLVDVGSQS PEEVVLSENL LHIAVRRKSV QMVRFLLRYK 780 PNKNSKGTAP MHLFRPDALG PSTITPLHIA ASTGDAEDVL DALTDDPGLV GISAWKHAQD 840 GTGFTPEDYA RQRGNDAYLN LVQKKIDKHL SKGHVVLGVP SSMCPVTNDG VKPGNVSLEI 900 CRSMPMAKCH LCSRQALKYP RSAVRTFLYR PAMLTMMGVA VVCVCVGILL HTMPKVYAAP 960 TFRWELLERG AM |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 4e-31 | 145 | 225 | 4 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3'. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00633 | PBM | Transfer from PK06791.1 | Download |
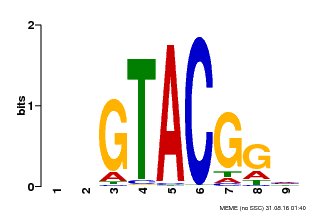
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 462951446 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002466159.1 | 0.0 | squamosa promoter-binding-like protein 6 | ||||
| Swissprot | Q75LH6 | 0.0 | SPL6_ORYSJ; Squamosa promoter-binding-like protein 6 | ||||
| TrEMBL | A0A0A9REY0 | 0.0 | A0A0A9REY0_ARUDO; Uncharacterized protein | ||||
| STRING | Sb01g002530.1 | 0.0 | (Sorghum bicolor) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1998 | 38 | 92 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G47070.1 | 0.0 | squamosa promoter binding protein-like 1 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



