 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 472338 | ||||||||
| Common Name | ARALYDRAFT_472338 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Arabidopsis
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 1041aa MW: 115334 Da PI: 8.2521 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 129.9 | 9.2e-41 | 126 | 203 | 1 | 78 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
+Cqv++C++dls+ak+yhrrhkvCevhska+++lv +++qrfCqqCsrfh lsefDe+krsCrrrLa+hn+rrrk+++
472338 126 MCQVDNCTEDLSHAKDYHRRHKVCEVHSKATKALVGKQMQRFCQQCSRFHLLSEFDEGKRSCRRRLAGHNRRRRKTTQ 203
6**************************************************************************875 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 7.6E-34 | 119 | 188 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 32.077 | 124 | 201 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 3.27E-37 | 125 | 203 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 9.2E-29 | 127 | 200 | IPR004333 | Transcription factor, SBP-box |
| CDD | cd00204 | 1.06E-8 | 791 | 934 | No hit | No description |
| Gene3D | G3DSA:1.25.40.20 | 2.0E-7 | 791 | 942 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 6.46E-8 | 832 | 938 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 9.092 | 834 | 947 | IPR020683 | Ankyrin repeat-containing domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005886 | Cellular Component | plasma membrane | ||||
| GO:0016021 | Cellular Component | integral component of membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1041 aa Download sequence Send to blast |
MDEVGAQVAA PMFIHHPMGK KRDLYYPMSN RLVQSQPRSD EWNSKMWDWD SRRFEAKPVD 60 VEVLRLGNEA QEFDLTLRNR SGEERGLDLN LGSGLTAVED LTTTTTQNGR PSKKVRSGSP 120 GGNYPMCQVD NCTEDLSHAK DYHRRHKVCE VHSKATKALV GKQMQRFCQQ CSRFHLLSEF 180 DEGKRSCRRR LAGHNRRRRK TTQPEEVASG VVVPGNRDNN NNTSTTNMDL MALLTALACA 240 QGKNAVKPAG SPAVPDREQL LQILNKINAL PLPMDLVSKL NNIGSLARKN MDHPTVNPQN 300 DMNGASPSTM DLLAVLSTTL GSSSPDALAI LSQGGFGNKD SEKTKLSSYE HGVTTNLEKR 360 TFGFSSVGGE RSSSSNQSPS QDSDSRGQDT RSSLSLQLFT SSPEDESRPT VASSRKYYSS 420 ASSNPAEDRS PSSSPVMQEL FPLQTSPETM RSKNHNNTSP RTGCLPLELF GASNRGAANP 480 NFKGFRQQSG YASSGSDYSP PSLNSDAQDR TGKIVFKLLD KDPSQLPGTL RSEIYNWLSN 540 IPSEMESYIR PGCVVLSVYV AMSPAAWEQL EQNLLQRLGV LLQNSSSDFW RNARFIVNTG 600 RQLASHKNGK VRCSKSWRTW NSPELISVSP VAVVAGEETS LVVRGRSLTN DGISIRCTHM 660 GSYMSMDVTG AVCRQAIFDK LNVDSFKVQN VHPGFLGRCF IEVENGFRGD SFPLIIANES 720 ICNELNRLEE EFHPKSQDMT EEPAQSSNRG PTSREEVLCF LNELGWLFQK NQTSEPREQS 780 DFSLTRFKFL LVCSVERDYC ALIRTLLDML VERNLVNDEL NREALEMLAE IQLLNRAVKR 840 KSTKMVELLI HYSVNPSALS SFKKFVFLPN RTGPGGITPL HIAACTSGSD DMIDLLTNDP 900 QEIGLSSWNT LCDATGQTPY SYAAMRNNHN YNSLVARKLA DKRNRQVSLN IENEIVDQTG 960 LSKRLSSEMN KSSTCASCAT VALKYQRRVS GSHRLFPTPI IHSMLAVATV CVCVCVFMHA 1020 FPIVRQGSHF SWGGLDYGSI * |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 5e-31 | 117 | 200 | 3 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3' (By similarity). May play a role in plant development. {ECO:0000250, ECO:0000269|PubMed:15703061}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00155 | DAP | Transfer from AT1G20980 | Download |
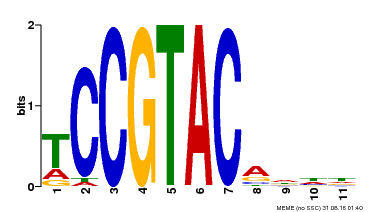
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 472338 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK226907 | 0.0 | AK226907.1 Arabidopsis thaliana mRNA for squamosa promoter binding protein-like 14, clone: RAFL09-16-H09. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020868897.1 | 0.0 | squamosa promoter-binding-like protein 14 | ||||
| Swissprot | Q8RY95 | 0.0 | SPL14_ARATH; Squamosa promoter-binding-like protein 14 | ||||
| TrEMBL | D7KJX2 | 0.0 | D7KJX2_ARALL; Uncharacterized protein | ||||
| STRING | fgenesh2_kg.1__2292__AT1G20980.1 | 0.0 | (Arabidopsis lyrata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM3999 | 25 | 54 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G20980.1 | 0.0 | squamosa promoter binding protein-like 14 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | 472338 |
| Entrez Gene | 9326482 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



