 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 473414 | ||||||||
| Common Name | ARALYDRAFT_473414 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Arabidopsis
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 388aa MW: 43728.4 Da PI: 8.2373 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 101.9 | 3.9e-32 | 213 | 266 | 2 | 55 |
G2-like 2 prlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
pr+rWt++LH+rFv+ave LGG+e+AtPk++lelm+vk+Ltl+hvkSHLQ+YR+
473414 213 PRMRWTTTLHARFVHAVELLGGHERATPKSVLELMDVKDLTLAHVKSHLQMYRT 266
9****************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.97E-15 | 210 | 266 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 2.0E-28 | 211 | 266 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 2.5E-23 | 213 | 266 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 2.6E-7 | 214 | 265 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009944 | Biological Process | polarity specification of adaxial/abaxial axis | ||||
| GO:0010158 | Biological Process | abaxial cell fate specification | ||||
| GO:0010229 | Biological Process | inflorescence development | ||||
| GO:0048481 | Biological Process | plant ovule development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 388 aa Download sequence Send to blast |
MELFPAQPDL SLQISPPNSK PTSTWQRRRS TTDQEDHEEL DLGFWRRALD SRTSSLVSNS 60 SSKTINHPFQ DLSLSNNSHH QQQQHHHHHP QLLPNCNGSN ILTSFQFPTQ QQQQHLQGFL 120 ARDLNTHLRP IRGIPLYHNP PPHHHRPPPP CFPFDPSSLI PSSSSSSPAL TGNNNSFNTS 180 SVSNPNYHNH HHQTLNRARF MPRFPAKRSM RAPRMRWTTT LHARFVHAVE LLGGHERATP 240 KSVLELMDVK DLTLAHVKSH LQMYRTVKTT DKAAASSGQS DVYENGSSGD NNSDDWMFDM 300 NRKSRDSEEL TNPLEKSNGL WTNSSGEARL HGKLIDNVAE IMLPSEKELD GKCSSYERIS 360 SEEMSSSSIS GTSPFKPNLE FTLGRSH* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4k_A | 2e-16 | 214 | 268 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4k_B | 2e-16 | 214 | 268 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_A | 2e-16 | 214 | 268 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_C | 2e-16 | 214 | 268 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_D | 2e-16 | 214 | 268 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_F | 2e-16 | 214 | 268 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_H | 2e-16 | 214 | 268 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_J | 2e-16 | 214 | 268 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that regulates lateral organ polarity. Promotes abaxial cell fate during lateral organd formation. Functions with KAN1 in the specification of polarity of the ovule outer integument. {ECO:0000269|PubMed:11525739, ECO:0000269|PubMed:15286295, ECO:0000269|PubMed:16623911, ECO:0000269|PubMed:17307928}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00174 | DAP | Transfer from AT1G32240 | Download |
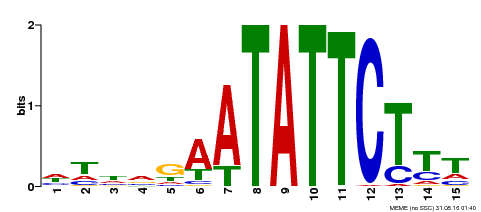
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 473414 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY048689 | 0.0 | AY048689.1 Arabidopsis thaliana GARP-like putative transcription factor KANADI2 (KAN2) mRNA, complete cds. | |||
| GenBank | BT029211 | 0.0 | BT029211.1 Arabidopsis thaliana At1g32240 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020870009.1 | 0.0 | probable transcription factor KAN2 | ||||
| Swissprot | Q9C616 | 0.0 | KAN2_ARATH; Probable transcription factor KAN2 | ||||
| TrEMBL | D7KHH9 | 0.0 | D7KHH9_ARALL; Uncharacterized protein | ||||
| STRING | fgenesh2_kg.1__3368__AT1G32240.1 | 0.0 | (Arabidopsis lyrata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM5387 | 28 | 50 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G32240.1 | 1e-179 | G2-like family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | 473414 |
| Entrez Gene | 9327038 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



