 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 476620 | ||||||||
| Common Name | ARALYDRAFT_476620 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Arabidopsis
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 258aa MW: 28540.9 Da PI: 7.5762 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 43.5 | 7.2e-14 | 97 | 141 | 3 | 47 |
SS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
+WT+eE++l++ + ++ G+g+Wk I+r + k Rt q+ s+ qky
476620 97 PWTEEEHKLFLLGLQRVGKGDWKGISRNFVKSRTSTQVASHAQKY 141
8*******************************************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 16.891 | 90 | 146 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 7.33E-17 | 92 | 147 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 4.5E-17 | 93 | 144 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 1.2E-9 | 94 | 144 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.6E-10 | 96 | 141 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 1.4E-10 | 97 | 141 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.32E-8 | 97 | 142 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009739 | Biological Process | response to gibberellin | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 258 aa Download sequence Send to blast |
MADGSTSSSE STNACSGGGT RREIMLFGVR VVLDPMRKCV SLNNLSEYEQ TAETPKIDGE 60 DRDGQDMNKT PAGYASADEA LPISSSNVNR ERKRGVPWTE EEHKLFLLGL QRVGKGDWKG 120 ISRNFVKSRT STQVASHAQK YFIRRSNLNR RRRRSSLFDM TTDTVNLQSE DDQVLMQENT 180 SQLSSPVPEI NNFSIHPVMQ VFPEFPVPTG NQSYGQLTSS NLIKLVPLTF QSSPAPLSLN 240 LSLASSNLNE PSPSMHSA |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Binds selectively to the DNA sequence 5'-[GA]GATAA-3' and may act as a transcription factor involved in the regulation of drought-responsive genes. Enhances stomatal closure in response to abscisic acid (ABA). Confers drought and salt tolerance. {ECO:0000269|PubMed:21030505}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00236 | DAP | Transfer from AT1G74840 | Download |
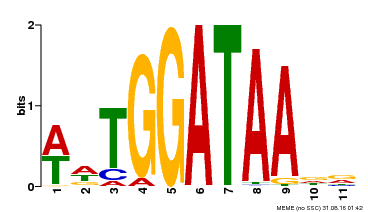
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 476620 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Up-regulated by drought, high salinity, and ABA. {ECO:0000269|PubMed:21030505}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY070098 | 0.0 | AY070098.1 Arabidopsis thaliana putative myb-related transcription activator protein (At1g74840) mRNA, complete cds. | |||
| GenBank | AY096393 | 0.0 | AY096393.1 Arabidopsis thaliana putative myb-related transcription activator (At1g74840) mRNA, complete cds. | |||
| GenBank | AY519510 | 0.0 | AY519510.1 Arabidopsis thaliana MYB transcription factor (At1g74840) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020891423.1 | 1e-164 | transcription factor MYB1R1 | ||||
| Swissprot | Q2V9B0 | 2e-54 | MY1R1_SOLTU; Transcription factor MYB1R1 | ||||
| TrEMBL | D7KS56 | 0.0 | D7KS56_ARALL; Myb family transcription factor (Fragment) | ||||
| STRING | fgenesh2_kg.2__1750__AT1G74840.1 | 0.0 | (Arabidopsis lyrata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM2461 | 26 | 69 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G74840.1 | 1e-138 | MYB_related family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | 476620 |
| Entrez Gene | 9325057 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



