 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 479964 | ||||||||
| Common Name | ARALYDRAFT_479964 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Arabidopsis
|
||||||||
| Family | HSF | ||||||||
| Protein Properties | Length: 328aa MW: 37473.8 Da PI: 5.9545 | ||||||||
| Description | HSF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HSF_DNA-bind | 110 | 1.7e-34 | 21 | 113 | 2 | 103 |
HHHHHHHHHCTGGGTTTSEESSSSSEEEES-HHHHHHHTHHHHSTT--HHHHHHHHHHTTEEE---SSBTTTTXTTSEEEEESXXXXXXXXXXXXXX CS
HSF_DNA-bind 2 FlkklyeiledeelkeliswsengnsfvvldeeefakkvLpkyFkhsnfaSFvRQLnmYgFkkvkdeekkskskekiweFkhksFkkgkkellekik 98
F+ k+y++++d+ ++ li+w nsf+v+d+ +f++++Lp yFkh+nf+SFvRQLn+YgF+kv+ ++ weF++++F +g+k+ll++i
479964 21 FIVKTYQMVNDPLTDWLITWGPAHNSFIVVDPLDFSQRILPAYFKHNNFSSFVRQLNTYGFRKVDPDR---------WEFANEHFLRGQKHLLKNIA 108
9********************999*****************************************999.........******************** PP
XXXXX CS
HSF_DNA-bind 99 rkkse 103
r+k++
479964 109 RRKHA 113
**975 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00415 | 1.9E-53 | 17 | 110 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| SuperFamily | SSF46785 | 1.5E-32 | 18 | 110 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| Gene3D | G3DSA:1.10.10.10 | 9.9E-37 | 18 | 110 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| PRINTS | PR00056 | 9.5E-18 | 21 | 44 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| Pfam | PF00447 | 1.2E-29 | 21 | 110 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| PRINTS | PR00056 | 9.5E-18 | 59 | 71 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| PROSITE pattern | PS00434 | 0 | 60 | 84 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| PRINTS | PR00056 | 9.5E-18 | 72 | 84 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0006970 | Biological Process | response to osmotic stress | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 328 aa Download sequence Send to blast |
MEDDNGNNTN NNNNNNVIAP FIVKTYQMVN DPLTDWLITW GPAHNSFIVV DPLDFSQRIL 60 PAYFKHNNFS SFVRQLNTYG FRKVDPDRWE FANEHFLRGQ KHLLKNIARR KHARGMYGQD 120 LEDGEIVREI ERLKDEQREL EAEIQRMNQR IEATEKRPEQ MMAFLYKVVE DPDLLPRMML 180 EKERTKQQVS DKKKRRVTMS TVKAEEEEVE EDEGRVFRVI SSSTPSPSST ENLHRNHSPD 240 GWVVPMTQGQ FGNYETGLVA NSMLSNSTSS TSSSLTSTFS LPESINGGGG GGCGNIQGER 300 RYKETATFGG VVESNPPTTP PYPFSLFR |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5d5u_B | 3e-25 | 18 | 110 | 26 | 129 | Heat shock factor protein 1 |
| 5d5v_B | 3e-25 | 18 | 110 | 26 | 129 | Heat shock factor protein 1 |
| 5d5v_D | 3e-25 | 18 | 110 | 26 | 129 | Heat shock factor protein 1 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 191 | 195 | KKKRR |
| 2 | 191 | 196 | KKKRRV |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
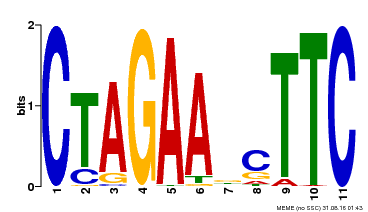
| UniProt | Transcriptional regulator that specifically binds DNA sequence 5'-AGAAnnTTCT-3' known as heat shock promoter elements (HSE). | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00379 | DAP | Transfer from AT3G24520 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 479964 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY056111 | 0.0 | AY056111.1 Arabidopsis thaliana AT3g24520/MOB24_5 mRNA, complete cds. | |||
| GenBank | AY072614 | 0.0 | AY072614.1 Arabidopsis thaliana AT3g24520/MOB24_5 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020889030.1 | 0.0 | heat stress transcription factor C-1 | ||||
| Swissprot | Q9LV52 | 0.0 | HSFC1_ARATH; Heat stress transcription factor C-1 | ||||
| TrEMBL | D7L548 | 0.0 | D7L548_ARALL; Uncharacterized protein (Fragment) | ||||
| STRING | fgenesh2_kg.3__2665__AT3G24520.1 | 0.0 | (Arabidopsis lyrata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM6775 | 27 | 43 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G24520.1 | 0.0 | heat shock transcription factor C1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | 479964 |
| Entrez Gene | 9321721 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



