 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 484115 | ||||||||
| Common Name | ARALYDRAFT_484115 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Arabidopsis
|
||||||||
| Family | BBR-BPC | ||||||||
| Protein Properties | Length: 281aa MW: 31330.8 Da PI: 10.2865 | ||||||||
| Description | BBR-BPC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | GAGA_bind | 361.5 | 1.7e-110 | 1 | 280 | 1 | 301 |
GAGA_bind 1 mdddgsrernkgyyepa.aslkenlglqlmssiaerdaki....rernlalsekkaavaerd..maflqrdkalaernkalverdnkllalllvenslas 93
mdddg+ rn+gyyepa as+k nlglqlms+i +r++k+ re+nl ++++++++++r+ m++ +++++ ++dnk++++l++++
484115 1 MDDDGF--RNWGYYEPAaASFKGNLGLQLMSTI-DRNTKPflpgRESNL-MIGSNGSYHPREhdMNY----SWISQ------PKDNKFFNMLPIST---- 82
99**99..9*******99***************.9**************.********988655999....*****......***********766.... PP
GAGA_bind 94 alpvgvqvlsgtksidslqqlsepqledsavelreeeklealpieeaaeeakekkkkkkrqrakkpkekkakkkkkksekskkkvkkesad.erskaekk 192
p++ +v+s+t+ +s+q++++p ++ s +e e+ + e ++ +k++k++ + ++++ pk+kk++k+k+++++ ++v + +r+k kk
484115 83 --PNYGSVMSETSGSNSMQMIHQPVVNSSRFE----EIPIPPREDEIVQPSKKRKMRGSISTPTIPKAKKMRKPKEERDVASSNV-----QqQRVKPAKK 171
..89999*************999888877777....3333344455666788888899999999999999999999554443333.....33779***** PP
GAGA_bind 193 sidlvlngvslDestlPvPvCsCtGalrqCYkWGnGGWqSaCCtttiSvyPLPvstkrrgaRiagrKmSqgafkklLekLaaeGydlsnpvDLkdhWAkH 292
s+dlv+ngvs+D+s+lPvPvC+CtG+++qCY+WG+GGWqSaCCtt+iSvyPLP+stkrrgaRi+grKmSqgafkk+LekLa+eGy+++n++DLk+hWA+H
484115 172 SVDLVINGVSMDISGLPVPVCTCTGTPQQCYRWGCGGWQSACCTTNISVYPLPMSTKRRGARISGRKMSQGAFKKVLEKLATEGYSFGNAIDLKSHWARH 271
**************************************************************************************************** PP
GAGA_bind 293 Gtnkfvtir 301
Gtnkfvtir
484115 272 GTNKFVTIR 280
********8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF06217 | 8.4E-100 | 1 | 280 | IPR010409 | GAGA-binding transcriptional activator |
| SMART | SM01226 | 9.0E-173 | 1 | 280 | IPR010409 | GAGA-binding transcriptional activator |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0050793 | Biological Process | regulation of developmental process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 281 aa Download sequence Send to blast |
MDDDGFRNWG YYEPAAASFK GNLGLQLMST IDRNTKPFLP GRESNLMIGS NGSYHPREHD 60 MNYSWISQPK DNKFFNMLPI STPNYGSVMS ETSGSNSMQM IHQPVVNSSR FEEIPIPPRE 120 DEIVQPSKKR KMRGSISTPT IPKAKKMRKP KEERDVASSN VQQQRVKPAK KSVDLVINGV 180 SMDISGLPVP VCTCTGTPQQ CYRWGCGGWQ SACCTTNISV YPLPMSTKRR GARISGRKMS 240 QGAFKKVLEK LATEGYSFGN AIDLKSHWAR HGTNKFVTIR * |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional regulator that specifically binds to GA-rich elements (GAGA-repeats) present in regulatory sequences of genes involved in developmental processes. Negatively regulates the homeotic gene AGL11/STK, which controls ovule primordium identity, by a cooperative binding to purine-rich elements present in the regulatory sequence leading to DNA conformational changes. {ECO:0000269|PubMed:14731261, ECO:0000269|PubMed:15722463}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00253 | DAP | Transfer from AT2G01930 | Download |
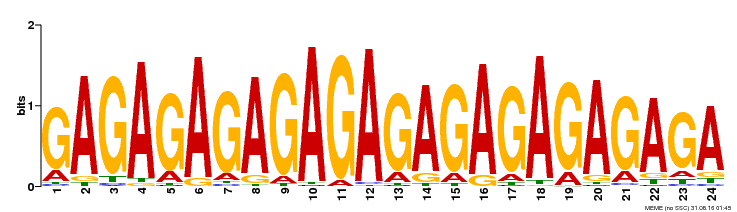
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 484115 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AC007265 | 0.0 | AC007265.5 Arabidopsis thaliana chromosome 2 clone F23I14 map CIC11A04, complete sequence. | |||
| GenBank | AY065225 | 0.0 | AY065225.1 Arabidopsis thaliana unknown protein (At2g01930) mRNA, complete cds. | |||
| GenBank | AY096550 | 0.0 | AY096550.1 Arabidopsis thaliana unknown protein (At2g01930) mRNA, complete cds. | |||
| GenBank | CP002685 | 0.0 | CP002685.1 Arabidopsis thaliana chromosome 2, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020880017.1 | 0.0 | protein BASIC PENTACYSTEINE1 | ||||
| Refseq | XP_020880018.1 | 0.0 | protein BASIC PENTACYSTEINE1 | ||||
| Swissprot | Q9SKD0 | 0.0 | BPC1_ARATH; Protein BASIC PENTACYSTEINE1 | ||||
| TrEMBL | D7LPX4 | 0.0 | D7LPX4_ARALL; Basic pentacysteine 1 | ||||
| STRING | fgenesh2_kg.5__107__AT2G01930.1 | 0.0 | (Arabidopsis lyrata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM2893 | 27 | 69 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G01930.2 | 0.0 | basic pentacysteine1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | 484115 |
| Entrez Gene | 9311169 |



