 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 484568 | ||||||||
| Common Name | ARALYDRAFT_484568 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Arabidopsis
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 434aa MW: 48879.4 Da PI: 7.2868 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 62.3 | 1e-19 | 184 | 230 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g WT+eEd+llv++v+++G++ W+ Ia++++ gR +kqc++rw+++l
484568 184 KGQWTPEEDKLLVQLVELHGTKKWSQIAKMLQ-GRVGKQCRERWHNHL 230
799*****************************.*************97 PP
| |||||||
| 2 | Myb_DNA-binding | 55.6 | 1.2e-17 | 236 | 278 | 1 | 45 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwq 45
++ WT+eEd +l++a+k+ G++ W+ Iar ++ gRt++ +k++w+
484568 236 KDVWTEEEDMILIKAHKEIGNR-WAEIARNLP-GRTENTIKNHWN 278
578*******************.*********.***********8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 30.906 | 179 | 234 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 2.39E-32 | 181 | 277 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 7.1E-18 | 183 | 232 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 2.8E-19 | 184 | 230 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.6E-29 | 185 | 238 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 5.23E-16 | 187 | 230 | No hit | No description |
| SMART | SM00717 | 5.4E-17 | 235 | 283 | IPR001005 | SANT/Myb domain |
| PROSITE profile | PS51294 | 20.744 | 235 | 285 | IPR017930 | Myb domain |
| Pfam | PF00249 | 1.9E-15 | 236 | 278 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 8.41E-13 | 239 | 281 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 4.0E-21 | 239 | 283 | IPR009057 | Homeodomain-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010439 | Biological Process | regulation of glucosinolate biosynthetic process | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:1904095 | Biological Process | negative regulation of endosperm development | ||||
| GO:2000692 | Biological Process | negative regulation of seed maturation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 434 aa Download sequence Send to blast |
MEFESVIKMH YPYLAAVMYD DSLALKDFHP SLTDDFSCVH NVHHKPSIPH TNEIPSKETI 60 RGITPSPCTE AFEAYFHGTS NDQGFFGMAY TTSPTIDESN VSHVPHDNTM WENDQSQGSV 120 FGTESTFNLA MVDSKPILSA NEDTIMNRRQ NNQVMIRTEQ ILKKNKRLQM RRICKPAKKA 180 SIIKGQWTPE EDKLLVQLVE LHGTKKWSQI AKMLQGRVGK QCRERWHNHL RPDIKKDVWT 240 EEEDMILIKA HKEIGNRWAE IARNLPGRTE NTIKNHWNAT KRRQHSRRTK GKDEISLALG 300 SNTLQNYIRS VTYNEDTFMT ANSNANIGPK NMRDKGKNVM VAVSKYEEDE CKYIVDGVMN 360 LGLDNGRIKM PSLAAVSAAS GSASTSGSAS GSGSGVTMEL DEPMTDSYMV MHGCDEVMMN 420 EIALLEMIAH GRL* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1gv2_A | 8e-45 | 182 | 284 | 2 | 104 | MYB PROTO-ONCOGENE PROTEIN |
| 1mse_C | 8e-45 | 182 | 284 | 2 | 104 | C-Myb DNA-Binding Domain |
| 1msf_C | 8e-45 | 182 | 284 | 2 | 104 | C-Myb DNA-Binding Domain |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 161 | 178 | LKKNKRLQMRRICKPAKK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that recognizes the motif 5'-TAACGG-3' in the promoter of endosperm-induced genes (PubMed:27681170, PubMed:25194028, PubMed:19066902). Promotes vegetative-to-embryonic transition and the formation of somatic embryos from root explants in a WUS-independent manner but via the expression of embryonic genes (e.g. LEC1, LEC2, FUS3 and WUS) (PubMed:18695688). May play an important role during embryogenesis and seed maturation (PubMed:19066902, PubMed:25194028). Together with MYB115, activates the transcription of S-ACP-DES2/AAD2 and S-ACP-DES3/AAD3 thus promoting the biosynthesis of omega-7 monounsaturated fatty acid in seed endosperm (PubMed:27681170). Regulates negatively maturation genes in the endosperm (PubMed:25194028). {ECO:0000269|PubMed:18695688, ECO:0000269|PubMed:19066902, ECO:0000269|PubMed:25194028, ECO:0000269|PubMed:27681170}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00385 | DAP | Transfer from AT3G27785 | Download |
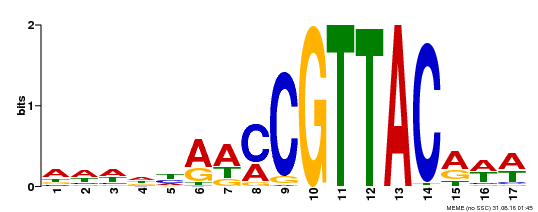
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 484568 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by LEC2. {ECO:0000269|PubMed:25194028}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AF334817 | 0.0 | AF334817.2 Arabidopsis thaliana putative transcription factor (MYB118) mRNA, complete cds. | |||
| GenBank | AY550306 | 0.0 | AY550306.1 Arabidopsis thaliana MYB transcription factor (At3g27785) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020880228.1 | 0.0 | transcription factor MYB118 isoform X2 | ||||
| Swissprot | Q9LVW4 | 0.0 | MY118_ARATH; Transcription factor MYB118 | ||||
| TrEMBL | D7LPK3 | 0.0 | D7LPK3_ARALL; MYB118 | ||||
| STRING | fgenesh2_kg.5__560__AT3G27785.1 | 0.0 | (Arabidopsis lyrata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM6052 | 26 | 43 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G27785.1 | 0.0 | myb domain protein 118 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | 484568 |
| Entrez Gene | 9313140 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



