 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 485204 | ||||||||
| Common Name | ARALYDRAFT_485204 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Arabidopsis
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 1358aa MW: 152229 Da PI: 7.2982 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 12.4 | 0.00048 | 1240 | 1265 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt.H 23
y+C C++sFs+ +L+ H r+ +
485204 1240 YQCDmeGCTMSFSSEKQLTLHKRNiC 1265
99********************9877 PP
| |||||||
| 2 | zf-C2H2 | 15.1 | 6.7e-05 | 1265 | 1287 | 3 | 23 |
ET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 3 Cp..dCgksFsrksnLkrHirtH 23
Cp Cgk F ++ +L++H+r+H
485204 1265 CPvkGCGKNFFSHKYLVQHQRVH 1287
9999*****************99 PP
| |||||||
| 3 | zf-C2H2 | 12 | 0.00065 | 1323 | 1349 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH..T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt..H 23
y+C Cg++F+ s++ rH r+ H
485204 1323 YVCAepGCGQTFRFVSDFSRHKRKtgH 1349
899999****************99666 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00545 | 7.6E-18 | 19 | 60 | IPR003349 | JmjN domain |
| PROSITE profile | PS51183 | 13.744 | 20 | 61 | IPR003349 | JmjN domain |
| Pfam | PF02375 | 6.3E-14 | 21 | 54 | IPR003349 | JmjN domain |
| SMART | SM00558 | 6.2E-53 | 200 | 369 | IPR003347 | JmjC domain |
| PROSITE profile | PS51184 | 33.99 | 203 | 369 | IPR003347 | JmjC domain |
| SuperFamily | SSF51197 | 8.79E-28 | 216 | 387 | No hit | No description |
| Pfam | PF02373 | 1.4E-37 | 233 | 352 | IPR003347 | JmjC domain |
| SMART | SM00355 | 7.5 | 1240 | 1262 | IPR015880 | Zinc finger, C2H2-like |
| SMART | SM00355 | 0.01 | 1263 | 1287 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 13.173 | 1263 | 1292 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 1.4E-6 | 1265 | 1291 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE pattern | PS00028 | 0 | 1265 | 1287 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 3.82E-10 | 1279 | 1321 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 9.6E-9 | 1292 | 1316 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.0015 | 1293 | 1317 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 10.928 | 1293 | 1322 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 1295 | 1317 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 3.04E-8 | 1311 | 1345 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 1.9E-9 | 1317 | 1346 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.62 | 1323 | 1349 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 11.032 | 1323 | 1354 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 1325 | 1349 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009741 | Biological Process | response to brassinosteroid | ||||
| GO:0009826 | Biological Process | unidimensional cell growth | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0033169 | Biological Process | histone H3-K9 demethylation | ||||
| GO:0035067 | Biological Process | negative regulation of histone acetylation | ||||
| GO:0048366 | Biological Process | leaf development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003676 | Molecular Function | nucleic acid binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1358 aa Download sequence Send to blast |
MAVSEQSQDV FPWLKSLPVA PEFRPTLAEF QDPIAYILKI EEEASRYGIC KILPPLPPPS 60 KKTSISNLNR SLAARAAARV RDGGFGACDY DGGPTFATRQ QQIGFCPRKQ RPVQRPVWQS 120 GEEYSFGEFE FKAKNFEKNY LKKCGKKSQL SALEIETLYW RATVDKPFSV EYANDMPGSA 180 FIPLSLAAAR RRESGGEGGT VGETAWNMRA MSRAEGSLLK FMKEEIPGVT SPMVYIAMMF 240 SWFAWHVEDH DLHSLNYLHM GAGKTWYGVP KDAAVAFEEV VRVHGYGGEL NPLVTFSTLG 300 EKTTVMSPEV FVKAGIPCCR LVQNPGDFVV TFPGAYHSGF SHGFNFGEAS NIATPEWLRM 360 AKDAAIRRAA INYPPMVSHL QLLYDFALAL GSRVPTSINA KPRSSRLKDK LRSEGERLTK 420 KLFVQNIIHN NELLSSLGKG SPVALLPQSS SDISVCSDLR IGSHLITNQE NPILLKSEDL 480 SSDSVMVGLS NGLKDTVSVK EKFTSLCERS RNHLASREKD TQETLSDAER RKNDGAVALS 540 DQRLFSCVTC GVLSFDCVAI VQPKEAAARY LMSADCSFFN DWTAASGSAN LGQAARSLHP 600 QSTEKHDVNY FYNDPVQTMD HSMKTGDQRT STTSLTMAHK DNGALGLLAS AYGDSSDSEE 660 EGQKGLDVPT SEGETKKYDQ EDADGNEEAR DGRTSDFNSQ RLTSEQNRLS KGGNSSLLEI 720 ALPFIPRSDD DSCRLHVFCL EHAAEVEQQL RPFGGIHIML LCHPEYPRIE AEAKIVAEEL 780 VINHEWNDTE FRNVTREDEE TIQAALDNVE PKGGNSDWTV KLGINLSYSA ILSRSPLYSK 840 QMPYNSVIYN AFGRSSPATS SPLKPEVSGK RSSRQRKYVV GKWCGKVWMS HQVHPFLLEQ 900 DLEGEESERN CHLRGALVED VTRTGLLPCN VSRDATTMFG RKYCRKRKVR AKAVPRKKLT 960 SFKREDGVSD DTSEDHSYKQ QWRASGNEEE SYFETGNTVS GDSSNQMSDQ QLKGIRRHRG 1020 VKEFESDNEV SDRSLGEEYT VRGCAASESS MENGFQQSMY DDDDDDDDID RHPRGIPRSE 1080 RIAVFRNPVS YDSEENGIYH QRGRVSRSNR QANRMGGEYD SEENSLEEQD FCSTGKRQTR 1140 STAKRKVKIE TVRSPRDTKG RTLQEFASGK KNEELDSYME GPSTRLRVRN LKPLRAASET 1200 KPKKIGKKRS GNASFSRVAT EEDMEEDEEA ENEEEECAAY QCDMEGCTMS FSSEKQLTLH 1260 KRNICPVKGC GKNFFSHKYL VQHQRVHSDD RPLKCPWKGC KMTFKWAWSR TEHIRVHTGA 1320 RPYVCAEPGC GQTFRFVSDF SRHKRKTGHS VKKTKKR* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6a57_A | 4e-79 | 1237 | 1357 | 20 | 140 | Lysine-specific demethylase REF6 |
| 6a58_A | 4e-79 | 1237 | 1357 | 20 | 140 | Lysine-specific demethylase REF6 |
| 6a59_A | 4e-79 | 1237 | 1357 | 20 | 140 | Lysine-specific demethylase REF6 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 945 | 957 | KRKVRAKAVPRKK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Histone demethylase that demethylates 'Lys-27' (H3K27me) of histone H3. Demethylates both tri- (H3K27me3) and di-methylated (H3K27me2) H3K27me (PubMed:21642989, PubMed:27111035). Demethylates also H3K4me3/2 and H3K36me3/2 in an in vitro assay (PubMed:20711170). Involved in the transcriptional regulation of hundreds of genes regulating developmental patterning and responses to various stimuli (PubMed:18467490). Binds DNA via its four zinc fingers in a sequence-specific manner, 5'-CTCTGYTY-3', to promote the demethylation of H3K27me3 and the regulation of organ boundary formation (PubMed:27111034, PubMed:27111035). Involved in the regulation of flowering time by repressing FLOWERING LOCUS C (FLC) expression (PubMed:15377760). Interacts with the NF-Y complexe to regulate SOC1 (PubMed:25105952). Mediates the recruitment of BRM to its target loci (PubMed:27111034). {ECO:0000269|PubMed:15377760, ECO:0000269|PubMed:18467490, ECO:0000269|PubMed:20711170, ECO:0000269|PubMed:21642989, ECO:0000269|PubMed:25105952, ECO:0000269|PubMed:27111034, ECO:0000269|PubMed:27111035}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00608 | ChIP-seq | Transfer from AT3G48430 | Download |
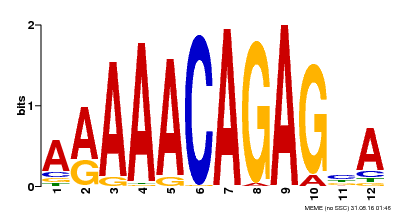
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 485204 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY664499 | 0.0 | AY664499.1 Arabidopsis thaliana relative of early flowering 6 (REF6) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002875902.1 | 0.0 | lysine-specific demethylase REF6 isoform X2 | ||||
| Swissprot | Q9STM3 | 0.0 | REF6_ARATH; Lysine-specific demethylase REF6 | ||||
| TrEMBL | D7LRW5 | 0.0 | D7LRW5_ARALL; Uncharacterized protein | ||||
| STRING | fgenesh2_kg.5__1196__AT3G48430.1 | 0.0 | (Arabidopsis lyrata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM5259 | 28 | 43 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G48430.1 | 0.0 | relative of early flowering 6 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | 485204 |
| Entrez Gene | 9311970 |



