 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 496013 | ||||||||
| Common Name | ARALYDRAFT_496013 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Arabidopsis
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 287aa MW: 31255.8 Da PI: 6.6309 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 45.9 | 1.3e-14 | 140 | 184 | 3 | 47 |
SS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
+WT+eE++l++ + k++G+g+W+ I+r + +Rt+ q+ s+ qky
496013 140 PWTEEEHKLFLMGLKKYGKGDWRNISRNFVITRTPTQVASHAQKY 184
8*****************************99************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51293 | 7.371 | 29 | 84 | IPR017884 | SANT domain |
| SMART | SM00717 | 1.9E-7 | 30 | 82 | IPR001005 | SANT/Myb domain |
| SuperFamily | SSF46689 | 5.02E-10 | 33 | 86 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 3.67E-7 | 33 | 79 | No hit | No description |
| PROSITE profile | PS51294 | 20.772 | 133 | 189 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 7.18E-17 | 135 | 188 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.8E-17 | 136 | 188 | IPR006447 | Myb domain, plants |
| Gene3D | G3DSA:1.10.10.60 | 1.3E-12 | 137 | 184 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 3.2E-13 | 137 | 187 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 4.9E-13 | 140 | 184 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.16E-11 | 140 | 185 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 287 aa Download sequence Send to blast |
MEVLRPTTSH VSGGNWLMDE TKSAVAASGE GATWTAAENK AFENALAVYD DNTPDRWQKV 60 AAVIPGKTVS DVIRQYNDLE ADVSSIEAGL IPVPGYITSP PFTLDWAGGG CNGFNPGHQV 120 CNKRSPAGRS PELERKKGVP WTEEEHKLFL MGLKKYGKGD WRNISRNFVI TRTPTQVASH 180 AQKYFIRQLP GGKDKRRASI HDITTVNLED EASLETSKSS IVVGEQRSRL TAFPWNPTDN 240 NGTHADAFNI TIGNAISGVH SYGQVLLGGF NNADSCYDAQ NTMFQL* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2cjj_A | 5e-18 | 26 | 99 | 3 | 76 | RADIALIS |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Involved in the dorsovental asymmetry of flowers. Promotes ventral identity. {ECO:0000269|PubMed:11937495, ECO:0000269|PubMed:9118809}. | |||||
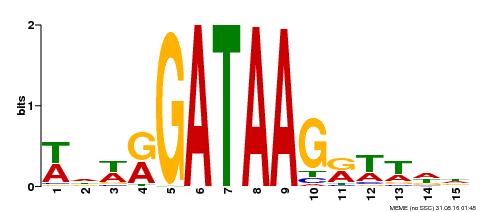
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00568 | DAP | Transfer from AT5G58900 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 496013 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK118891 | 0.0 | AK118891.1 Arabidopsis thaliana At5g58900 mRNA for putative I-box binding factor, complete cds, clone: RAFL21-22-M07. | |||
| GenBank | AY519533 | 0.0 | AY519533.1 Arabidopsis thaliana MYB transcription factor (At5g58900) mRNA, complete cds. | |||
| GenBank | BT005473 | 0.0 | BT005473.1 Arabidopsis thaliana clone U51211 putative I-box binding factor (At5g58900) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020869590.1 | 0.0 | transcription factor DIVARICATA | ||||
| Swissprot | Q8S9H7 | 1e-103 | DIV_ANTMA; Transcription factor DIVARICATA | ||||
| TrEMBL | D7MR44 | 0.0 | D7MR44_ARALL; Myb family transcription factor | ||||
| STRING | fgenesh2_kg.8__1829__AT5G58900.1 | 0.0 | (Arabidopsis lyrata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM1228 | 27 | 100 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G58900.1 | 0.0 | Homeodomain-like transcriptional regulator | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | 496013 |
| Entrez Gene | 9300675 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



