 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 51845 | ||||||||
| Common Name | MICPUCDRAFT_51845 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Chlorophyta; prasinophytes; Mamiellophyceae; Mamiellales; Mamiellaceae; Micromonas; Micromonas pusilla
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 746aa MW: 75641.9 Da PI: 9.8045 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 52 | 1.6e-16 | 98 | 142 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
r rWT eE+e+++ a k++G W++I +++g ++++ q++s+ qk+
51845 98 RERWTDEEHERFLAALKLHGRA-WRKIEEHVG-TKSAVQIRSHAQKF 142
78******************88.*********.************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 7.48E-16 | 92 | 146 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 21.549 | 93 | 147 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.5E-8 | 95 | 145 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.1E-14 | 96 | 145 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 8.8E-13 | 97 | 145 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 4.3E-13 | 98 | 141 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 2.33E-9 | 100 | 143 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0007623 | Biological Process | circadian rhythm | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009734 | Biological Process | auxin-activated signaling pathway | ||||
| GO:0010600 | Biological Process | regulation of auxin biosynthetic process | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 746 aa Download sequence Send to blast |
MSNPPPAWPG YAQAAAASGG FGPGVARIKP TPPVQQGALP ASNAAPWARG QTPPGGDATS 60 RGLGALSNAA TSNATQHAVA NVGHVPKARK PYTITKQRER WTDEEHERFL AALKLHGRAW 120 RKIEEHVGTK SAVQIRSHAQ KFFSKLMREA AKSGDASGVA SAGVSGSASE HGVSASVIPP 180 ARPKRKPAHP YPRKAPEGGD RAKNATTMSG DRRTVSTDGG TRTEVSDGTI TGADVNSFAA 240 FGHGAFGHGG FDAAHAAALA SSYAQFPAAP AAGPAAGATN AASYFNPYLL PANPTAAMYQ 300 MNAALKGATN ATTPRAANAT TDDAAQAAAM RRFMHAFQGL TGMPAAMTAP AAPPPPAPQP 360 GPPQPGPAAH GGGGAAAAGA NPSAATDFFA AMMAAATSNA SANLGSAESA FSQWAAAAAA 420 QQQSNTAQAQ QAAAMSQMMM MMMPGYAQAA AAVATAAGGA AAANPNPNPN QNPNADADAD 480 ANANLQSLAD FAVKQHRPRE FEEPQPVAEK KPEFDFNAVA ARKARSTRAH PKAAAAARAS 540 AEPPFPPAST AANDDASDPA SEGDLNKESS PENDGSRPSS HERGGASGSG EEDERDATAA 600 ARGGGGGETA SHTTPFAWCT PFLKDFSRRH SSPALPFQRL TGGGGGATTD AKTINTSGSN 660 ESTQLHADGE RGANHHRGRG GAGANAGAGK SQDNFELCAD MSSRRELSPS SEEGGEDHNQ 720 KSHSRPPPAA RLKLSTQTRN ARSRA* |
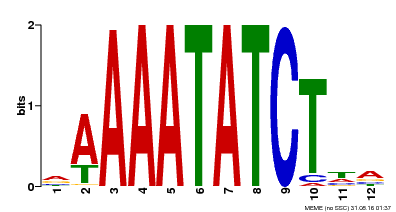
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00146 | DAP | Transfer from AT1G18330 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KR089140 | 5e-53 | KR089140.1 Micromonas pusilla isolate 1545_2 NADH dehydrogenase gene, partial cds. | |||
| GenBank | KR089201 | 5e-53 | KR089201.1 Micromonas pusilla isolate 1545_1 NADH dehydrogenase gene, partial cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_003062228.1 | 0.0 | predicted protein | ||||
| TrEMBL | C1N246 | 0.0 | C1N246_MICPC; Predicted protein | ||||
| STRING | XP_003062228.1 | 0.0 | (Micromonas pusilla) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Chlorophytae | OGCP454 | 14 | 27 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G17300.1 | 1e-31 | MYB_related family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | 51845 |
| Entrez Gene | 9687467 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



