 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 55361 | ||||||||
| Common Name | MICPUN_55361 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Chlorophyta; prasinophytes; Mamiellophyceae; Mamiellales; Mamiellaceae; Micromonas
|
||||||||
| Family | E2F/DP | ||||||||
| Protein Properties | Length: 284aa MW: 31421.4 Da PI: 4.8602 | ||||||||
| Description | E2F/DP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | E2F_TDP | 74.1 | 1.4e-23 | 17 | 74 | 8 | 70 |
E2F_TDP 8 lltqkflkllekseegivtlnevakeLvsedvknkrRRiYDilNVLealnliekkekneirwk 70
llt+kf++l+ ++e+g+++ln++a+ L +v ++RRiYDi+NVLe+++liekk+kn+i wk
55361 17 LLTKKFISLIDRAEHGTIDLNQAAEVL---KV--QKRRIYDITNVLEGIGLIEKKSKNNILWK 74
79*************************...88..****************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.10 | 4.0E-23 | 9 | 74 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| SMART | SM01372 | 1.0E-25 | 10 | 75 | IPR003316 | E2F/DP family, winged-helix DNA-binding domain |
| Pfam | PF02319 | 9.9E-22 | 17 | 74 | IPR003316 | E2F/DP family, winged-helix DNA-binding domain |
| SuperFamily | SSF46785 | 1.59E-15 | 17 | 73 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| SuperFamily | SSF144074 | 1.26E-16 | 88 | 154 | No hit | No description |
| Pfam | PF16421 | 3.7E-19 | 89 | 153 | IPR032198 | E2F transcription factor, CC-MB domain |
| CDD | cd14660 | 1.46E-27 | 89 | 154 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0051446 | Biological Process | positive regulation of meiotic cell cycle | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005667 | Cellular Component | transcription factor complex | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 284 aa Download sequence Send to blast |
MHASGTTPCR YDRLSPLLTK KFISLIDRAE HGTIDLNQAA EVLKVQKRRI YDITNVLEGI 60 GLIEKKSKNN ILWKPSASAP AFPEANIMKG NLYIAEEDIK NIPSFSSDTL VAVRAPYGTT 120 LEVPDPDEGD ELSKKRYQIL LKSSSGPVDV FLVSLQGNNG THGPKDSSRQ RKTCLHGQFS 180 KTSDEILLNK QEPGETGSTQ FETGSDAVVL NAHDIFHLET AEASGLKERM FDSPLVADET 240 GFKTPNMLRI VPPPGDQDYW FLQDARDMNM GLQDLFASYD FED* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1cf7_A | 3e-21 | 7 | 74 | 4 | 72 | PROTEIN (TRANSCRIPTION FACTOR E2F-4) |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 42 | 48 | LKVQKRR |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds DNA cooperatively with DP proteins through the E2 recognition site, 5'-TTTC[CG]CGC-3' found in the promoter region of a number of genes whose products are involved in cell cycle regulation or in DNA replication. The binding of retinoblastoma-related proteins represses transactivation. Regulates gene expression both positively and negatively. Activates the expression of E2FB. Involved in the control of cell-cycle progression from G1 to S phase. Stimulates cell proliferation and delays differentiation. {ECO:0000269|PubMed:11669580, ECO:0000269|PubMed:11786543, ECO:0000269|PubMed:11862494, ECO:0000269|PubMed:11889041, ECO:0000269|PubMed:11891240, ECO:0000269|PubMed:12913157, ECO:0000269|PubMed:15377755, ECO:0000269|PubMed:16514015, ECO:0000269|PubMed:19662336}. | |||||
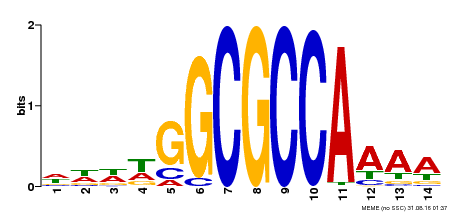
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00293 | DAP | Transfer from AT2G36010 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | CP001574 | 0.0 | CP001574.1 Micromonas sp. RCC299 chromosome 1, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002507264.1 | 0.0 | e2f1-like protein | ||||
| Swissprot | Q9FNY0 | 1e-45 | E2FA_ARATH; Transcription factor E2FA | ||||
| TrEMBL | C1FE93 | 0.0 | C1FE93_MICCC; E2f1-like protein | ||||
| STRING | XP_002507264.1 | 0.0 | (Micromonas sp. RCC299) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Chlorophytae | OGCP3258 | 13 | 14 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G36010.1 | 3e-48 | E2F transcription factor 3 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | 55361 |
| Entrez Gene | 8250587 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



