 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 59370 | ||||||||
| Common Name | MICPUN_59370 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Chlorophyta; prasinophytes; Mamiellophyceae; Mamiellales; Mamiellaceae; Micromonas
|
||||||||
| Family | WOX | ||||||||
| Protein Properties | Length: 191aa MW: 20595.2 Da PI: 8.2094 | ||||||||
| Description | WOX family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 50.2 | 4.3e-16 | 117 | 177 | 2 | 57 |
T--SS--HHHHHHHHHHHHH.SSS--HHHHHHHHHHC....TS-HHHHHHHHHHHHHHHHC CS
Homeobox 2 rkRttftkeqleeLeelFek.nrypsaeereeLAkkl....gLterqVkvWFqNrRakekk 57
R+t+++ ql +LeelF+ +p+ + r++++++l ++e++V++WFqN++a+ kk
59370 117 GARWTPSAAQLARLEELFATgMGTPNGDLRTKITDELaklgPVNEANVYNWFQNKKARTKK 177
68*****************99*****************9999*****************97 PP
| |||||||
| 2 | Wus_type_Homeobox | 76.8 | 3.1e-25 | 118 | 178 | 4 | 64 |
Wus_type_Homeobox 4 tRWtPtpeQikiLeelyksGlrtPnkeeiqritaeLeeyGkiedkNVfyWFQNrkaRerqk 64
+RWtP++ Q+ Leel+ +G+ tPn + ++it eL++ G ++++NV++WFQN+kaR ++k
59370 118 ARWTPSAAQLARLEELFATGMGTPNGDLRTKITDELAKLGPVNEANVYNWFQNKKARTKKK 178
7**********************************************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50071 | 11.969 | 113 | 178 | IPR001356 | Homeobox domain |
| SuperFamily | SSF46689 | 5.01E-12 | 114 | 178 | IPR009057 | Homeodomain-like |
| SMART | SM00389 | 9.9E-8 | 115 | 182 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 2.45E-7 | 116 | 178 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 3.8E-11 | 116 | 184 | IPR009057 | Homeodomain-like |
| Pfam | PF00046 | 1.1E-13 | 117 | 177 | IPR001356 | Homeobox domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0001944 | Biological Process | vasculature development | ||||
| GO:0010072 | Biological Process | primary shoot apical meristem specification | ||||
| GO:0051301 | Biological Process | cell division | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 191 aa Download sequence Send to blast |
MSEGLCANDP IFDVPPLPTD EQVKRFGMDG DGPYPTIAPG AMTWTQLDTL RHQIAAFSHL 60 CEQRALLTQQ IYEESKPGHQ RSQTLAAIAN SIGVSAAGSQ PSYPGPVYSK GGDGPRGARW 120 TPSAAQLARL EELFATGMGT PNGDLRTKIT DELAKLGPVN EANVYNWFQN KKARTKKKLL 180 EEQAAKNRSG * |
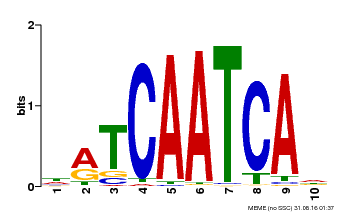
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00617 | PBM | Transfer from AT4G35550 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | CP001327 | 0.0 | CP001327.1 Micromonas sp. RCC299 chromosome 6, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002503252.1 | 1e-141 | predicted protein | ||||
| TrEMBL | C1E8G9 | 1e-139 | C1E8G9_MICCC; Uncharacterized protein | ||||
| STRING | XP_002503252.1 | 1e-140 | (Micromonas sp. RCC299) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Chlorophytae | OGCP6445 | 6 | 7 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G20700.1 | 2e-20 | WUSCHEL related homeobox 14 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | 59370 |
| Entrez Gene | 8244181 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



