 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 59779 | ||||||||
| Common Name | MICPUN_59779 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Chlorophyta; prasinophytes; Mamiellophyceae; Mamiellales; Mamiellaceae; Micromonas
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 1565aa MW: 163846 Da PI: 6.212 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 161.5 | 1.5e-50 | 55 | 171 | 2 | 117 |
CG-1 2 lkekkrwlkneeiaaiLenfekheltl..elktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptfqrrcywlLe 103
++++rwlkn+e++++L n++++ ++l +++ rp+ g+++L++rk vr+frkDG+ w+kkkdgktvrE+hekLKvg+ve+l+cyYah+ en fqrrcywlL+
59779 55 NQSRTRWLKNTEVCDMLLNYRSYGFALskTAPVRPPAGTIFLFDRKAVRFFRKDGHDWQKKKDGKTVRETHEKLKVGNVELLNCYYAHAAENDRFQRRCYWLLD 158
3459******************9988766899************************************************************************ PP
CG-1 104 eelekivlvhylev 117
++ e +vlvhyl++
59779 159 SD-EGVVLVHYLDT 171
**.********987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 67.269 | 50 | 177 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 8.1E-57 | 53 | 172 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 8.7E-49 | 57 | 170 | IPR005559 | CG-1 DNA-binding domain |
| SuperFamily | SSF81296 | 1.66E-14 | 694 | 780 | IPR014756 | Immunoglobulin E-set |
| CDD | cd00204 | 2.93E-12 | 982 | 1074 | No hit | No description |
| SuperFamily | SSF48403 | 2.8E-15 | 982 | 1076 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 3.4E-15 | 983 | 1077 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 16.759 | 985 | 1086 | IPR020683 | Ankyrin repeat-containing domain |
| Pfam | PF12796 | 4.6E-7 | 994 | 1075 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50088 | 11.915 | 1018 | 1050 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 4.4E-5 | 1018 | 1047 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50096 | 7.693 | 1500 | 1528 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0071275 | Biological Process | cellular response to aluminum ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1565 aa Download sequence Send to blast |
MYQPRVSPSY NAGGSDSPAT GSDGGFGGVS ATALIMPEAE GPDGSTRAHV IEMLNQSRTR 60 WLKNTEVCDM LLNYRSYGFA LSKTAPVRPP AGTIFLFDRK AVRFFRKDGH DWQKKKDGKT 120 VRETHEKLKV GNVELLNCYY AHAAENDRFQ RRCYWLLDSD EGVVLVHYLD TTMVVGGRGG 180 AKGAGLGLAP GGSAPGSNSG TTDGEDQNGF RNLFRYASGM SQYSGSGYEE VASMGGMSGG 240 SPMMQSPARP GQYHPEVYQQ YSQQQQAVGG QYGHHGYGAP PTHHYPHGDS DPHALYHAAQ 300 EQFNGVAESE SWFNSVLASE DEAGEESGPG GPGRMSGVHR RASATLVDAA GRAVPFRQPS 360 SGRAVPFREG SNRSILSIGS GPPLDHGYFE MGQDESSNGP HFDLLKEFAV DQIAEAQGIK 420 VEGMPVESAA AAAAAAAAAG PNIGTAFPGE YRSDIADGVT SMAELESKIA SLKETLVRVA 480 SVNADASPEQ VDRLEQDMAA LERGAAAMRP AAGSVGGGGG GAKPGHRRGS SFGSGTRARP 540 PRARPVIWDG DDKDEDDADR EDDGDDDDDD GDDNEETAEG ALSPLAKAAA ATRRPRGADG 600 GRGAHRAAAL AAQQQQRMAG FPRVPSHPSE GSEMDAEMNN SDGDNGDISP RAPSPALRAA 660 QMRAAAEASH ARYNRARQPF IPPAQQGSPI LWAIDDFSPE WDTETGGGKV LVTGTPRPGL 720 PEGLYLCCVF GDVEVPAEQV SPGVLRCRAP PMNAGRVPFY ISCLGSGKRP ASDIRTFEYR 780 EAGAGGARDK RAAEIRLTSG VTERDFQLRL VHLLIGADRS SKASGGGGPG GPGGPGGGGG 840 GGGGGSTASG NGPSDGKGSG GSGGNTSSPD NRNGGGSSES RDVHGSPASP GRSGSQSGGL 900 GGGAGAGFTR RAVAALNLGA DPALDPNMSN LSDEDVGKVF KTALEARLRH AIAAEAKHHR 960 ARAEGIVPNP TYVQPKSAYA RVDAGGMGLI HCVAALGMKW AIPAMTKCGC DVNQPDRRNR 1020 TALHWAAAKG HEDTVATLLA SGANIRAMAR WGAGGYTAAD LAAALGHGGI AAYISETSLA 1080 ASLSNISLYG GAQGPATGRS LRSASFDRKR QQQQQQVQGL TTPLMSGAGV TPPGVTPGAG 1140 PTANRTGRAQ RDRVKAVPMR SLLLATETEV TATDAVTSQT EVETEAEFME GGGTSTERRE 1200 AVAAGMIQLA FRKHSVRRRK ARKKGGELNT VTEGVETGAA PRTGADRDVK KDVKKVVKKA 1260 EKAAQKITDS LRSLKVSKTA TGAAPELGKR RLGERSGGGL HHSTIPESEA MDVSVEEAAG 1320 SPPEGAEGAD FAMRSTSGGE EALAALRRRI AQLQERAVAL QGEIRAADYR GRQDRLQTLM 1380 PGGHHQRRRA ADIMQLISAG VGGGGGGGGG GEDGAAAARV VDWERAMLER ANSSKDSLDE 1440 VYRSGGGASN RLVKSATLRG RRQSHKSHSD DGNDSNKRST SDTNNNDDDS DDDEEEENVQ 1500 QAVARIQAIT RSREARDQYL RLRMVTQSMQ ERRDAIARGE AVKREEDEDG LDDVDVEEDE 1560 EEDE* |
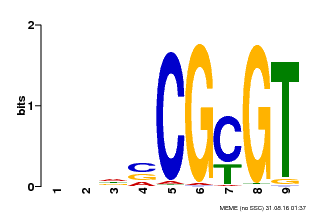
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00043 | PBM | Transfer from AT5G64220 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | CP001328 | 0.0 | CP001328.1 Micromonas sp. RCC299 chromosome 7, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002503449.1 | 0.0 | camta-like transcriptional regulator | ||||
| TrEMBL | C1E9N0 | 0.0 | C1E9N0_MICCC; Camta-like transcriptional regulator | ||||
| STRING | XP_002503449.1 | 0.0 | (Micromonas sp. RCC299) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Chlorophytae | OGCP5132 | 8 | 9 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G64220.2 | 3e-42 | Calmodulin-binding transcription activator protein with CG-1 and Ankyrin domains | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | 59779 |
| Entrez Gene | 8244740 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



