 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 60948 | ||||||||
| Common Name | MICPUN_60948 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Chlorophyta; prasinophytes; Mamiellophyceae; Mamiellales; Mamiellaceae; Micromonas
|
||||||||
| Family | E2F/DP | ||||||||
| Protein Properties | Length: 823aa MW: 81033.4 Da PI: 6.2193 | ||||||||
| Description | E2F/DP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | E2F_TDP | 74 | 1.6e-23 | 130 | 195 | 1 | 71 |
E2F_TDP 1 rkeksLrlltqkflkllekseegivtlnevakeLvsedvknkrRRiYDilNVLealnliekkekneirwkg 71
rk+ksL+ll+++fl+l+ +e+ ++l+e+a +L v +rRRiYDi+NVLe+++++ +k+kn+++w+g
60948 130 RKDKSLGLLCENFLHLYGAGQEELISLDEAAAKL---GV--ERRRIYDIVNVLESVEVVVRKAKNKYTWHG 195
79********************************...**..****************************98 PP
| |||||||
| 2 | E2F_TDP | 70.2 | 2.5e-22 | 352 | 432 | 1 | 71 |
E2F_TDP 1 rkeksLrlltqkflkllekseegivtlnevakeLvse.....dvknkrRRiYDilNVLealnliek.....kekneirwkg 71
r+eksL+ll+qkf++l+ s++++v+l+++a++L+ + k+k+RR+YDi+N+L++l+liek ++k+++rw g
60948 352 RREKSLGLLSQKFVQLFLVSRARVVSLESAARTLLGScadqaKLKTKVRRLYDIANILSSLRLIEKthlvdSRKPAFRWLG 432
689*******************************999999**99*************************999*******87 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.10 | 1.3E-26 | 127 | 196 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| SuperFamily | SSF46785 | 2.27E-15 | 129 | 194 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| SMART | SM01372 | 9.6E-30 | 130 | 195 | IPR003316 | E2F/DP family, winged-helix DNA-binding domain |
| Pfam | PF02319 | 1.1E-20 | 131 | 195 | IPR003316 | E2F/DP family, winged-helix DNA-binding domain |
| Gene3D | G3DSA:1.10.10.10 | 2.8E-23 | 349 | 434 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| SMART | SM01372 | 4.7E-30 | 352 | 432 | IPR003316 | E2F/DP family, winged-helix DNA-binding domain |
| SuperFamily | SSF46785 | 1.5E-11 | 354 | 433 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| Pfam | PF02319 | 3.1E-19 | 354 | 432 | IPR003316 | E2F/DP family, winged-helix DNA-binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0032876 | Biological Process | negative regulation of DNA endoreduplication | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005667 | Cellular Component | transcription factor complex | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 823 aa Download sequence Send to blast |
MQAPAAGGSS AAAAAPQGTA AAQPQPVIPG VAPAHAGAPL AGDPMAAALA AAQQFLGFMS 60 HAGVNPGGFP PGPAPGLFAP PGAPQGPARP GSAAAMFSPD HHQMRPGEGP ASRGATPGVG 120 GEMAGATYNR KDKSLGLLCE NFLHLYGAGQ EELISLDEAA AKLGVERRRI YDIVNVLESV 180 EVVVRKAKNK YTWHGISRMP AALDRLWKEG KREFGEDLVL DGGDDDKDSP RGKAKIAELE 240 AERAKVKAEV NGDHGSAAKP EIDEGDDDED DVDDKSEGVK SEGDAESGGS GDAKSSGGSN 300 KDANKDAKSE EEDEDAAVAV AALTGENGAA SSADKSSGAK PSGAGGGTGD CRREKSLGLL 360 SQKFVQLFLV SRARVVSLES AARTLLGSCA DQAKLKTKVR RLYDIANILS SLRLIEKTHL 420 VDSRKPAFRW LGVEKELAAL QAAAAAGVKA EPGHPAAATA AAAAHAFAAH ATSGSSGAPA 480 TGAPAFNPQW FMPNVGGRAM TPPVPRVGSP FDAAASMAAA AAAANFAASV GVPERPGSSL 540 GKSKRGGGAS GGGGSPSRSD PSRSDPASDP GEIKRPRSAL GDAARSHLSA PTPQRAVAPG 600 LDALSAMAEI GSSGDVGVGG GSTGAGAAAL AAPVASRPLP GGFATAPSPA SSPGYAQMMA 660 AMAAAGIASP APAPDAAAQQ AATLAMLASM GPGGAEMMQA MFQQQQAQAQ SQAQAMMAAA 720 AAASAAGAPG AGGAPAAAAA AQAAQAYYQA MAAASFKPPP PGAGPAAPTP GAAPAGPGAL 780 PHGVVGPMGD YMEMWRKWSA QYAGGAPPGA PSASAPEANG AQ* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4yo2_A | 2e-18 | 129 | 424 | 3 | 228 | Transcription factor E2F8 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00397 | DAP | Transfer from AT3G48160 | Download |
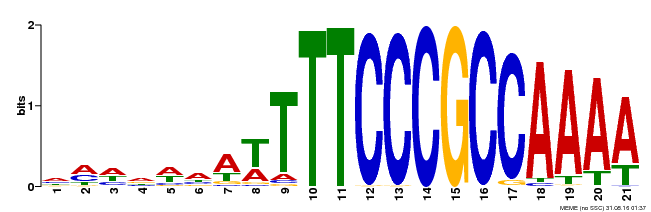
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | CP001575 | 0.0 | CP001575.1 Micromonas sp. RCC299 chromosome 8, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002508040.1 | 0.0 | predicted protein | ||||
| TrEMBL | C1FGP3 | 0.0 | C1FGP3_MICCC; Uncharacterized protein | ||||
| STRING | XP_002508040.1 | 0.0 | (Micromonas sp. RCC299) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Chlorophytae | OGCP4186 | 10 | 12 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G48160.1 | 5e-29 | DP-E2F-like 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | 60948 |
| Entrez Gene | 8245950 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



