 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 67638 | ||||||||
| Common Name | SELMODRAFT_67638 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Lycopodiidae; Selaginellales; Selaginellaceae; Selaginella
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 171aa MW: 19600.2 Da PI: 9.8308 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 105.5 | 6.8e-33 | 10 | 148 | 1 | 128 |
NAM 1 lppGfrFhPtdeelvveyLkkkvegkk.....leleev.ikev.diykvePwdLpkkvkaeekewyfFskrdkkyatgkrknratk.....sgyWkatgkdkev 92
lp+G++F+P+d+e++ ++L++k+ + ++++ +++ +i++ +P++Lp vk++ ++ +fF++ +++y++g+rk+r+++ + +W++tgk+++v
67638 10 LPAGVKFDPNDKEIL-DHLAAKIGRGGkphalIDEFIPtLEDDeGICSKHPEKLP-GVKSDGSSSHFFHRPSNAYTKGTRKRRKVQtddddETRWHKTGKTRPV 111
799***********9.89999998766565443332222333336**********.77888999*******************75545666899********** PP
NAM 93 lskkgelvglkktLvfykg..rapkgektdWvmheyrl 128
++ +ge++g kk++v+y++ +++k++kt+Wv+h+y++
67638 112 ME-NGEVIGWKKIMVLYTNfkKKSKPKKTNWVIHQYHV 148
**.999***********6433888899*********97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 3.27E-35 | 4 | 167 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 30.615 | 10 | 167 | IPR003441 | NAC domain |
| Pfam | PF02365 | 3.4E-14 | 11 | 148 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 171 aa Download sequence Send to blast |
SQAPQDWTGL PAGVKFDPND KEILDHLAAK IGRGGKPHAL IDEFIPTLED DEGICSKHPE 60 KLPGVKSDGS SSHFFHRPSN AYTKGTRKRR KVQTDDDDET RWHKTGKTRP VMENGEVIGW 120 KKIMVLYTNF KKKSKPKKTN WVIHQYHVGI HEDEREGELV MSKVFFQTQP R |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator involved in xylem formation (PubMed:25265867, PubMed:25535195). Promotes the expression of the secondary wall-associated transcription factor MYB46 (PubMed:25265867). Functions upstream of NAC030/VND7, a master switch of xylem vessel differentiation (PubMed:25265867, PubMed:25535195). Acts as upstream regulator of NAC101/VND6 and LBD30/ASL19 (PubMed:25535195). {ECO:0000269|PubMed:25265867, ECO:0000269|PubMed:25535195}. | |||||
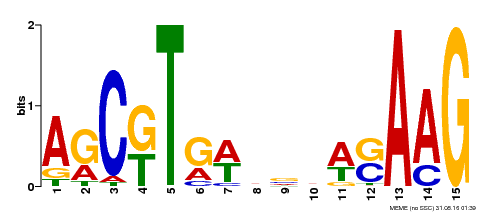
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00460 | DAP | Transfer from AT4G29230 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024536647.1 | 1e-124 | NAC domain-containing protein 82 isoform X1 | ||||
| Refseq | XP_024536648.1 | 1e-124 | NAC domain-containing protein 73 isoform X3 | ||||
| Swissprot | Q9M0F8 | 5e-82 | NAC75_ARATH; NAC domain-containing protein 75 | ||||
| TrEMBL | D8RBR5 | 1e-124 | D8RBR5_SELML; Uncharacterized protein (Fragment) | ||||
| STRING | EFJ30836 | 1e-125 | (Selaginella moellendorffii) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Representative plant | OGRP525 | 15 | 85 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G29230.1 | 2e-80 | NAC domain containing protein 75 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | 67638 |
| Entrez Gene | 9631469 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



